+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6k2k | ||||||
|---|---|---|---|---|---|---|---|
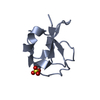
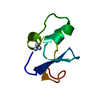
| Title | Solution structure of MUL1-RING domain | ||||||
 Components Components | Mitochondrial ubiquitin ligase activator of NFKB 1 | ||||||
 Keywords Keywords | STRUCTURAL PROTEIN / Solution structure / E3 ubiquitin ligase | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway / TOM complex / negative regulation of chemokine (C-C motif) ligand 5 production / negative regulation of defense response to virus by host / negative regulation of mitochondrial fusion / positive regulation of dendrite extension / positive regulation of protein sumoylation / positive regulation of type 2 mitophagy / mitochondrion localization / mitochondrial fission ...regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway / TOM complex / negative regulation of chemokine (C-C motif) ligand 5 production / negative regulation of defense response to virus by host / negative regulation of mitochondrial fusion / positive regulation of dendrite extension / positive regulation of protein sumoylation / positive regulation of type 2 mitophagy / mitochondrion localization / mitochondrial fission / regulation of mitochondrion organization / SUMO transferase activity / negative regulation of type I interferon-mediated signaling pathway / positive regulation of mitochondrial fission / cellular response to exogenous dsRNA / protein sumoylation / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / negative regulation of innate immune response / regulation of mitochondrial membrane potential / RING-type E3 ubiquitin transferase / protein destabilization / protein polyubiquitination / p53 binding / ubiquitin-protein transferase activity / ubiquitin protein ligase activity / KEAP1-NFE2L2 pathway / peroxisome / Neddylation / mitochondrial outer membrane / positive regulation of canonical NF-kappaB signal transduction / Ub-specific processing proteases / protein stabilization / protein ubiquitination / axon / neuronal cell body / apoptotic process / ubiquitin protein ligase binding / mitochondrion / zinc ion binding / identical protein binding / membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Lee, M.S. / Lee, M.K. / Ryu, K.S. / Chi, S.W. | ||||||
 Citation Citation |  Journal: Biochem.Biophys.Res.Commun. / Year: 2019 Journal: Biochem.Biophys.Res.Commun. / Year: 2019Title: Solution structure of MUL1-RING domain and its interaction with p53 transactivation domain. Authors: Lee, M.S. / Lee, S.O. / Lee, M.K. / Yi, G.S. / Lee, C.K. / Ryu, K.S. / Chi, S.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6k2k.cif.gz 6k2k.cif.gz | 384.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6k2k.ent.gz pdb6k2k.ent.gz | 323.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6k2k.json.gz 6k2k.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k2/6k2k https://data.pdbj.org/pub/pdb/validation_reports/k2/6k2k ftp://data.pdbj.org/pub/pdb/validation_reports/k2/6k2k ftp://data.pdbj.org/pub/pdb/validation_reports/k2/6k2k | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 6251.479 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: MUL1, C1orf166, GIDE, MAPL, MULAN, RNF218 / Plasmid: pGEX-4T3 / Production host: Homo sapiens (human) / Gene: MUL1, C1orf166, GIDE, MAPL, MULAN, RNF218 / Plasmid: pGEX-4T3 / Production host:  References: UniProt: Q969V5, RING-type E3 ubiquitin transferase |
|---|---|
| #2: Chemical |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Type: solution Contents: 0.9 mM [U-99% 13C; U-99% 15N] Mitochondrial ubiquitin ligase activator of NFKB 1, 90% H2O/10% D2O Details: 13C, 15N-labeled MUL1-RING domain / Label: 13C, 15N_sample / Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample | Conc.: 0.9 mM Component: Mitochondrial ubiquitin ligase activator of NFKB 1 Isotopic labeling: [U-99% 13C; U-99% 15N] |
| Sample conditions | Details: 50 mM MES, pH 6.5, 50 mM NaCl, 5 microM Zinc sulfate, 10 mM dithiothreitol (DTT), and 10 % (v/v) D2O Ionic strength: 50 mM / Label: conditions_1 / pH: 6.5 / Pressure: 1 atm / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Bruker AVANCE II / Manufacturer: Bruker / Model: AVANCE II / Field strength: 900 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 | ||||||||||||||||||
| NMR representative | Selection criteria: minimized average structure | ||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 250 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller










 PDBj
PDBj


 HSQC
HSQC