+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6jnm | ||||||
|---|---|---|---|---|---|---|---|
| Title | REF6 ZnF2-4-NAC004-mC3 complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA BINDING PROTEIN/DNA / REF6 / zinc finger / 5mC / DNA complex / DNA BINDING PROTEIN-DNA complex | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of ethylene-activated signaling pathway / abscisic acid catabolic process / release of seed from dormancy / positive regulation of lateral root development / response to diterpene / heat acclimation / protein localization to heterochromatin / sugar mediated signaling pathway / unidimensional cell growth / vegetative to reproductive phase transition of meristem ...regulation of ethylene-activated signaling pathway / abscisic acid catabolic process / release of seed from dormancy / positive regulation of lateral root development / response to diterpene / heat acclimation / protein localization to heterochromatin / sugar mediated signaling pathway / unidimensional cell growth / vegetative to reproductive phase transition of meristem / regulation of photoperiodism, flowering / systemic acquired resistance / response to brassinosteroid / histone H3K27me2/H3K27me3 demethylase activity / leaf development / ethylene-activated signaling pathway / Oxidoreductases; Acting on paired donors, with incorporation or reduction of molecular oxygen; With 2-oxoglutarate as one donor, and incorporation of one atom of oxygen into each donor / response to abscisic acid / Ino80 complex / abscisic acid-activated signaling pathway / response to mechanical stimulus / epigenetic regulation of gene expression / protein homooligomerization / response to heat / sequence-specific DNA binding / positive regulation of gene expression / regulation of DNA-templated transcription / protein homodimerization activity / zinc ion binding / nucleus Similarity search - Function | ||||||
| Biological species |  synthetic construct (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.05 Å MOLECULAR REPLACEMENT / Resolution: 2.05 Å | ||||||
 Authors Authors | Yao, Q.Q. / Wu, B.X. / Ma, J.B. | ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: DNA methylation repels targeting of Arabidopsis REF6. Authors: Qiu, Q. / Mei, H. / Deng, X. / He, K. / Wu, B. / Yao, Q. / Zhang, J. / Lu, F. / Ma, J. / Cao, X. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6jnm.cif.gz 6jnm.cif.gz | 85.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6jnm.ent.gz pdb6jnm.ent.gz | 60.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6jnm.json.gz 6jnm.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jn/6jnm https://data.pdbj.org/pub/pdb/validation_reports/jn/6jnm ftp://data.pdbj.org/pub/pdb/validation_reports/jn/6jnm ftp://data.pdbj.org/pub/pdb/validation_reports/jn/6jnm | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6jnlSC  6jnnC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 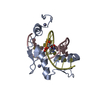
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 12026.109 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q9STM3, Oxidoreductases; Acting on paired donors, with incorporation or reduction of molecular oxygen; With 2-oxoglutarate as one donor, and incorporation of one atom of oxygen into each donor #2: DNA chain | Mass: 3639.387 Da / Num. of mol.: 2 / Source method: obtained synthetically / Details: 5CM / Source: (synth.) synthetic construct (others) #3: DNA chain | Mass: 3697.472 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) #4: Chemical | ChemComp-ZN / #5: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.66 Å3/Da / Density % sol: 66.38 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop Details: 16% PEG 3,350, 0.03 M citrate acid, 0.07 M Bis-Tris propane pH 7.6 |
-Data collection
| Diffraction | Mean temperature: 293 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRF SSRF  / Beamline: BL19U1 / Wavelength: 0.97853 Å / Beamline: BL19U1 / Wavelength: 0.97853 Å |
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: Jun 17, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97853 Å / Relative weight: 1 |
| Reflection | Resolution: 2.05→30 Å / Num. obs: 35297 / % possible obs: 100 % / Redundancy: 12.3 % / Rmerge(I) obs: 0.102 / Net I/σ(I): 4.241 |
| Reflection shell | Resolution: 2.05→2.12 Å / Rmerge(I) obs: 1.02 / Num. unique obs: 3546 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6JNL Resolution: 2.05→30 Å / Cor.coef. Fo:Fc: 0.943 / Cor.coef. Fo:Fc free: 0.924 / SU B: 3.905 / SU ML: 0.102 / Cross valid method: THROUGHOUT / ESU R: 0.152 / ESU R Free: 0.141 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 28.27 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.05→30 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj









































