Entry Database : PDB / ID : 6jijTitle The Crystal Structure of Main Protease from Mouse Hepatitis Virus A59 in Complex with an inhibitor 02J-ALA-VAL-LEU-PJE-010 Replicative polyprotein 1ab Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species synthetic construct (others) Method / / / Resolution : 2.65 Å Authors Cui, W. / Cui, S.S. Funding support Organization Grant number Country Ministry of Science and Technology (China) 2016YFD0500300 Ministry of Science and Technology (China) 2015CB859800 National Natural Science Foundation of China 31528006
Journal : Biochem. Biophys. Res. Commun. / Year : 2019Title : The crystal structure of main protease from mouse hepatitis virus A59 in complex with an inhibitor.Authors : Cui, W. / Cui, S. / Chen, C. / Chen, X. / Wang, Z. / Yang, H. / Zhang, L. History Deposition Feb 21, 2019 Deposition site / Processing site Revision 1.0 Apr 24, 2019 Provider / Type Revision 1.1 Feb 19, 2020 Group Category pdbx_struct_assembly / pdbx_struct_assembly_gen ... pdbx_struct_assembly / pdbx_struct_assembly_gen / pdbx_struct_oper_list / struct_site / struct_site_gen Item / _pdbx_struct_assembly_gen.asym_id_list / _pdbx_struct_assembly_gen.oper_expressionRevision 2.0 Nov 15, 2023 Group Atomic model / Data collection ... Atomic model / Data collection / Database references / Derived calculations Category atom_site / chem_comp_atom ... atom_site / chem_comp_atom / chem_comp_bond / database_2 / struct_conn Item _atom_site.auth_atom_id / _atom_site.label_atom_id ... _atom_site.auth_atom_id / _atom_site.label_atom_id / _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr2_label_atom_id Revision 2.1 Oct 23, 2024 Group / Category / pdbx_modification_feature
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Murine coronavirus
Murine coronavirus X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.65 Å
MOLECULAR REPLACEMENT / Resolution: 2.65 Å  Authors
Authors China, 3items
China, 3items  Citation
Citation Journal: Biochem. Biophys. Res. Commun. / Year: 2019
Journal: Biochem. Biophys. Res. Commun. / Year: 2019 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6jij.cif.gz
6jij.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6jij.ent.gz
pdb6jij.ent.gz PDB format
PDB format 6jij.json.gz
6jij.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/ji/6jij
https://data.pdbj.org/pub/pdb/validation_reports/ji/6jij ftp://data.pdbj.org/pub/pdb/validation_reports/ji/6jij
ftp://data.pdbj.org/pub/pdb/validation_reports/ji/6jij Links
Links Assembly
Assembly


 Components
Components Murine coronavirus (strain A59) / Strain: A59 / Production host:
Murine coronavirus (strain A59) / Strain: A59 / Production host: 
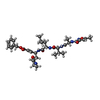
 Type: Peptide-like / Class: Inhibitor / Mass: 680.791 Da / Num. of mol.: 3 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others)
Type: Peptide-like / Class: Inhibitor / Mass: 680.791 Da / Num. of mol.: 3 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SSRF
SSRF  / Beamline: BL17B1 / Wavelength: 0.97854 Å
/ Beamline: BL17B1 / Wavelength: 0.97854 Å Processing
Processing MOLECULAR REPLACEMENT / Resolution: 2.65→36.96 Å / SU ML: 0.38 / Cross valid method: FREE R-VALUE / σ(F): 1.37 / Phase error: 31.25 / Stereochemistry target values: ML
MOLECULAR REPLACEMENT / Resolution: 2.65→36.96 Å / SU ML: 0.38 / Cross valid method: FREE R-VALUE / σ(F): 1.37 / Phase error: 31.25 / Stereochemistry target values: ML Movie
Movie Controller
Controller






















 PDBj
PDBj






