Entry Database : PDB / ID : 6j0eTitle Structures of two ArsR As(III)-responsive repressors: implications for the mechanism of derepression Arsenic responsive repressor ArsR Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / Biological species Corynebacterium glutamicum ATCC 13032 (bacteria)Method / / / Resolution : 1.6 Å Authors Prabaharan, C. / Kandavelu, P. / Packianathan, C. / Rosen, P.B. / Thiyagarajan, S. Funding support Organization Grant number Country Other government EMR/2014/000299 National Institutes of Health/National Library of Medicine (NIH/NLM) R01 GM55425 National Institutes of Health/National Library of Medicine (NIH/NLM) S10_RR25528 National Institutes of Health/National Library of Medicine (NIH/NLM) S10_RR028976
Journal : J.Struct.Biol. / Year : 2019Title : Structures of two ArsR As(III)-responsive transcriptional repressors: Implications for the mechanism of derepression.Authors : Prabaharan, C. / Kandavelu, P. / Packianathan, C. / Rosen, B.P. / Thiyagarajan, S. History Deposition Dec 24, 2018 Deposition site / Processing site Revision 1.0 Jul 3, 2019 Provider / Type Revision 1.1 Aug 7, 2019 Group / Database references / Category Item / _citation.page_first / _citation.page_lastRevision 1.2 Mar 23, 2022 Group / Database references / Category / pdbx_audit_supportItem / _database_2.pdbx_database_accession / _pdbx_audit_support.funding_organizationRevision 1.3 May 29, 2024 Group / Category / chem_comp_bond
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Corynebacterium glutamicum ATCC 13032 (bacteria)
Corynebacterium glutamicum ATCC 13032 (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MAD / Resolution: 1.6 Å
MAD / Resolution: 1.6 Å  Authors
Authors India,
India,  United States, 4items
United States, 4items  Citation
Citation Journal: J.Struct.Biol. / Year: 2019
Journal: J.Struct.Biol. / Year: 2019 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6j0e.cif.gz
6j0e.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6j0e.ent.gz
pdb6j0e.ent.gz PDB format
PDB format 6j0e.json.gz
6j0e.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/j0/6j0e
https://data.pdbj.org/pub/pdb/validation_reports/j0/6j0e ftp://data.pdbj.org/pub/pdb/validation_reports/j0/6j0e
ftp://data.pdbj.org/pub/pdb/validation_reports/j0/6j0e Links
Links Assembly
Assembly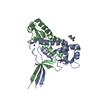
 Components
Components Corynebacterium glutamicum ATCC 13032 (bacteria)
Corynebacterium glutamicum ATCC 13032 (bacteria)
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  APS
APS  / Beamline: 22-ID / Wavelength: 1.04478, 1.00495
/ Beamline: 22-ID / Wavelength: 1.04478, 1.00495 Processing
Processing MAD / Resolution: 1.6→29.79 Å / Cor.coef. Fo:Fc: 0.949 / Cor.coef. Fo:Fc free: 0.901 / SU B: 4.806 / SU ML: 0.077 / Cross valid method: FREE R-VALUE / ESU R: 0.194 / ESU R Free: 0.136 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
MAD / Resolution: 1.6→29.79 Å / Cor.coef. Fo:Fc: 0.949 / Cor.coef. Fo:Fc free: 0.901 / SU B: 4.806 / SU ML: 0.077 / Cross valid method: FREE R-VALUE / ESU R: 0.194 / ESU R Free: 0.136 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS Movie
Movie Controller
Controller













 PDBj
PDBj



