[English] 日本語
 Yorodumi
Yorodumi- PDB-6ijw: SOLUTION NMR STRUCTURE OF A DODECAMERIC dsDNA COMPLEXED WITH A NI... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ijw | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
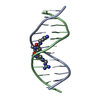
| Title | SOLUTION NMR STRUCTURE OF A DODECAMERIC dsDNA COMPLEXED WITH A NIR FLUORESCENT PROBE QCy-DT | ||||||||||||||||||||
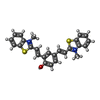
 Components Components | DNA (5'-D(* Keywords KeywordsDNA / dsDNA / ligand / QCy-DT | Function / homology | Chem-QCY / DNA / DNA (> 10) |  Function and homology information Function and homology informationBiological species |  Homo sapiens (human) Homo sapiens (human)Method | SOLUTION NMR / torsion angle dynamics |  Authors AuthorsGanguly, S. / Basu, G. |  Citation Citation Journal: Biochemistry / Year: 2021 Journal: Biochemistry / Year: 2021Title: DNA Minor Groove-Induced cis - trans Isomerization of a Near-Infrared Fluorescent Probe. Authors: Ganguly, S. / Murugan, N.A. / Ghosh, D. / Narayanaswamy, N. / Govindaraju, T. / Basu, G. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ijw.cif.gz 6ijw.cif.gz | 81.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ijw.ent.gz pdb6ijw.ent.gz | 55.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ijw.json.gz 6ijw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ij/6ijw https://data.pdbj.org/pub/pdb/validation_reports/ij/6ijw ftp://data.pdbj.org/pub/pdb/validation_reports/ij/6ijw ftp://data.pdbj.org/pub/pdb/validation_reports/ij/6ijw | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5zldC  6ijvC C: citing same article ( |
|---|---|
| Similar structure data | |
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 3663.392 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) Homo sapiens (human)#2: Chemical | ChemComp-QCY / | |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||
| Sample conditions | Ionic strength: 10mM NaCl mM / Label: conditions_1 / pH: 7 / Pressure: 1 atm / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Bruker AVANCE III / Manufacturer: Bruker / Model: AVANCE III / Field strength: 700 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics / Software ordinal: 5 | ||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 20 / Conformers submitted total number: 4 |
 Movie
Movie Controller
Controller









 PDBj
PDBj







































 CYANA
CYANA