[English] 日本語
 Yorodumi
Yorodumi- PDB-6hac: Crystal structure of [Fe]-hydrogenase (Hmd) holoenzyme from Metha... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6hac | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of [Fe]-hydrogenase (Hmd) holoenzyme from Methanococcus aeolicus (open form) | ||||||
 Components Components | 5,10-methenyltetrahydromethanopterin hydrogenase | ||||||
 Keywords Keywords | OXIDOREDUCTASE / [Fe]-hydrogenase / catalytic cycle / conformational rearrangement / Fe-guanylylpyridinol cofactor / methanogenesis / hydride-transfer / tetrahydromethanopterin / C1-metabolism | ||||||
| Function / homology |  Function and homology information Function and homology information5,10-methenyltetrahydromethanopterin hydrogenase / N5,N10-methenyltetrahydromethanopterin hydrogenase activity / methanogenesis, from carbon dioxide / pyrroline-5-carboxylate reductase activity / L-proline biosynthetic process / one-carbon metabolic process Similarity search - Function | ||||||
| Biological species | Methanococcus aeolicus | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.3 Å MOLECULAR REPLACEMENT / Resolution: 2.3 Å | ||||||
 Authors Authors | Huang, G. / Wagner, T. / Wodrich, M.D. / Ataka, K. / Bill, E. / Ermler, U. / Hu, X. / Shima, S. | ||||||
| Funding support |  Germany, 1items Germany, 1items
| ||||||
 Citation Citation |  Journal: Nat Catal / Year: 2019 Journal: Nat Catal / Year: 2019Title: The atomic-resolution crystal structure of activated [Fe]-hydrogenase Authors: Huang, G. / Wagner, T. / Wodrich, M.D. / Ataka, K. / Bill, E. / Ermler, U. / Hu, X. / Shima, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6hac.cif.gz 6hac.cif.gz | 150.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6hac.ent.gz pdb6hac.ent.gz | 116.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6hac.json.gz 6hac.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6hac_validation.pdf.gz 6hac_validation.pdf.gz | 747.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6hac_full_validation.pdf.gz 6hac_full_validation.pdf.gz | 750.1 KB | Display | |
| Data in XML |  6hac_validation.xml.gz 6hac_validation.xml.gz | 15.5 KB | Display | |
| Data in CIF |  6hac_validation.cif.gz 6hac_validation.cif.gz | 21.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ha/6hac https://data.pdbj.org/pub/pdb/validation_reports/ha/6hac ftp://data.pdbj.org/pub/pdb/validation_reports/ha/6hac ftp://data.pdbj.org/pub/pdb/validation_reports/ha/6hac | HTTPS FTP |
-Related structure data
| Related structure data |  6haeC  6havC  4jjfS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 36784.562 Da / Num. of mol.: 1 / Mutation: wild-type Source method: isolated from a genetically manipulated source Details: / Source: (gene. exp.)  Methanococcus aeolicus (strain ATCC BAA-1280 / DSM 17508 / OCM 812 / Nankai-3) (archaea) Methanococcus aeolicus (strain ATCC BAA-1280 / DSM 17508 / OCM 812 / Nankai-3) (archaea)Tissue: / / Cell: / / Cell line: / / Gene: hmd, Maeo_1025 / Organ: / / Variant: / / Plasmid: pET-24b+ / Details (production host): / / Cell (production host): / / Cell line (production host): / / Organ (production host): / / Production host:  References: UniProt: A6UVT1, 5,10-methenyltetrahydromethanopterin hydrogenase |
|---|
-Non-polymers , 5 types, 112 molecules 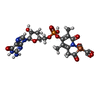








| #2: Chemical | ChemComp-FE9 / | ||||
|---|---|---|---|---|---|
| #3: Chemical | ChemComp-1PE / | ||||
| #4: Chemical | | #5: Chemical | ChemComp-NA / | #6: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.74 Å3/Da / Density % sol: 67.17 % / Description: Transparent diamond shape |
|---|---|
| Crystal grow | Temperature: 281.15 K / Method: vapor diffusion, sitting drop / pH: 4 Details: The reconstituted [Fe]-hydrogenase holoenzyme from M. aeolicus was crystallized in the anaerobic tent with gas phase 95%N2/5%H2 using 96-well 2-drop MRC Crystallization Plates (Molecular ...Details: The reconstituted [Fe]-hydrogenase holoenzyme from M. aeolicus was crystallized in the anaerobic tent with gas phase 95%N2/5%H2 using 96-well 2-drop MRC Crystallization Plates (Molecular Dimensions, Suffolk, UK). For the initial screening, 0.7-ul of 24 mg/ml reconstituted [Fe]-hydrogenase was mixed with 0.7 ul of reservoir solution of crystallization kits under yellow light and incubated under the dark condition. The best diffracting crystal was obtained in one month in the crystallization reservoir solution containing 20% w/v polyethylene glycol 3350, 100-mM tri-sodium citrate pH 4.0 and 200-mM tri-sodium citrate. PH range: / Temp details: Temperature variation was +/- 1 during crystallization |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: BM30A / Wavelength: 0.9798 Å / Beamline: BM30A / Wavelength: 0.9798 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Sep 26, 2016 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9798 Å / Relative weight: 1 |
| Reflection | Resolution: 2.3→48.43 Å / Num. obs: 25078 / % possible obs: 100 % / Redundancy: 13.7 % / Biso Wilson estimate: 39.33 Å2 / CC1/2: 0.999 / Rmerge(I) obs: 0.155 / Rpim(I) all: 0.043 / Rrim(I) all: 0.161 / Net I/σ(I): 17.9 |
| Reflection shell | Resolution: 2.3→2.42 Å / Redundancy: 14 % / Rmerge(I) obs: 1.023 / Mean I/σ(I) obs: 2.9 / Num. unique obs: 3607 / CC1/2: 0.602 / Rpim(I) all: 0.281 / Rrim(I) all: 1.061 / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4JJF Resolution: 2.3→26.62 Å / Cor.coef. Fo:Fc: 0.95 / Cor.coef. Fo:Fc free: 0.946 / SU R Cruickshank DPI: 0.351 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.177 / SU Rfree Blow DPI: 0.146 / SU Rfree Cruickshank DPI: 0.146
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 43.63 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.27 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 2.3→26.62 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.3→2.39 Å / Total num. of bins used: 13
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: -2.1917 Å / Origin y: -41.5428 Å / Origin z: -22.9285 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Selection details: { A|* } |
 Movie
Movie Controller
Controller












 PDBj
PDBj






