+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6fyv | ||||||
|---|---|---|---|---|---|---|---|
| Title | X-RAY STRUCTURE OF CLK4-KD(146-480)/CX-4945 AT 2.46A | ||||||
 Components Components | Dual specificity protein kinase CLK4 | ||||||
 Keywords Keywords | TRANSFERASE / SPLICING / KINASE / PHOSPHOTRANSFERASE | ||||||
| Function / homology |  Function and homology information Function and homology informationdual-specificity kinase / regulation of RNA splicing / protein serine/threonine/tyrosine kinase activity / protein tyrosine kinase activity / protein serine kinase activity / protein serine/threonine kinase activity / ATP binding / nucleus Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.46 Å molecular replacement / Resolution: 2.46 Å | ||||||
 Authors Authors | Kallen, J. | ||||||
 Citation Citation |  Journal: ChemMedChem / Year: 2018 Journal: ChemMedChem / Year: 2018Title: X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome. Authors: Kallen, J. / Bergsdorf, C. / Arnaud, B. / Bernhard, M. / Brichet, M. / Cobos-Correa, A. / Elhajouji, A. / Freuler, F. / Galimberti, I. / Guibourdenche, C. / Haenni, S. / Holzinger, S. / ...Authors: Kallen, J. / Bergsdorf, C. / Arnaud, B. / Bernhard, M. / Brichet, M. / Cobos-Correa, A. / Elhajouji, A. / Freuler, F. / Galimberti, I. / Guibourdenche, C. / Haenni, S. / Holzinger, S. / Hunziker, J. / Izaac, A. / Kaufmann, M. / Leder, L. / Martus, H.J. / von Matt, P. / Polyakov, V. / Roethlisberger, P. / Roma, G. / Stiefl, N. / Uteng, M. / Lerchner, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6fyv.cif.gz 6fyv.cif.gz | 86.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6fyv.ent.gz pdb6fyv.ent.gz | 63.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6fyv.json.gz 6fyv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6fyv_validation.pdf.gz 6fyv_validation.pdf.gz | 749.2 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6fyv_full_validation.pdf.gz 6fyv_full_validation.pdf.gz | 750.6 KB | Display | |
| Data in XML |  6fyv_validation.xml.gz 6fyv_validation.xml.gz | 14.9 KB | Display | |
| Data in CIF |  6fyv_validation.cif.gz 6fyv_validation.cif.gz | 20.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fy/6fyv https://data.pdbj.org/pub/pdb/validation_reports/fy/6fyv ftp://data.pdbj.org/pub/pdb/validation_reports/fy/6fyv ftp://data.pdbj.org/pub/pdb/validation_reports/fy/6fyv | HTTPS FTP |
-Related structure data
| Related structure data |  6fyiC  6fykC  6fylSC  6fyoC  6fypC  6fyrC C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 39691.496 Da / Num. of mol.: 1 / Fragment: kinase domain Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CLK4 / Production host: Homo sapiens (human) / Gene: CLK4 / Production host:  | ||
|---|---|---|---|
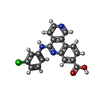
| #2: Chemical | ChemComp-3NG / | ||
| #3: Chemical | | #4: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.64 Å3/Da / Density % sol: 53.45 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 6.5 Details: 30% Pentaerythritol ethoxylate (15/4 EO/OH),0.05M Ammonium Sulfate, 0.05M TRIS |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X10SA / Wavelength: 1 Å / Beamline: X10SA / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Aug 5, 2016 |
| Radiation | Monochromator: SI 111 channel / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.46→19.36 Å / Num. obs: 16267 / % possible obs: 99.6 % / Redundancy: 12.9 % / Biso Wilson estimate: 55.7 Å2 / CC1/2: 1 / Rmerge(I) obs: 0.084 / Rrim(I) all: 0.087 / Net I/σ(I): 23.9 |
| Reflection shell | Resolution: 2.46→2.52 Å / Redundancy: 13.3 % / Rmerge(I) obs: 0.889 / Mean I/σ(I) obs: 3.4 / CC1/2: 0.895 / Rrim(I) all: 0.924 / % possible all: 100 |
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6FYL Resolution: 2.46→19.36 Å / Cor.coef. Fo:Fc: 0.952 / Cor.coef. Fo:Fc free: 0.952 / SU B: 7.581 / SU ML: 0.171 / SU R Cruickshank DPI: 0.4361 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.436 / ESU R Free: 0.242 Details: HYDROGENS HAVE BEEN USED IF PRESENT IN THE INPUT U VALUES : REFINED INDIVIDUALLY
| |||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | |||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 144.38 Å2 / Biso mean: 59.556 Å2 / Biso min: 29.36 Å2
| |||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.46→19.36 Å
| |||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.46→2.523 Å / Rfactor Rfree error: 0 / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller
















 PDBj
PDBj