+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6evh | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Lipoaminopeptide helioferin A and B from Mycogone rosea | |||||||||
 Components Components | Lipoaminopeptide helioferin A and B | |||||||||
 Keywords Keywords | ANTIBIOTIC / Lipoaminopeptide / protonophoric / peptaibol / peptaibiotic | |||||||||
| Function / homology | FLUORIDE ION Function and homology information Function and homology information | |||||||||
| Biological species |  Mycogone rosea (fungus) Mycogone rosea (fungus) | |||||||||
| Method |  X-RAY DIFFRACTION / AB INITIO PHASING / Resolution: 0.9 Å X-RAY DIFFRACTION / AB INITIO PHASING / Resolution: 0.9 Å | |||||||||
 Authors Authors | Gessmann, R. / Petratos, K. | |||||||||
 Citation Citation |  Journal: Acta Cryst. D / Year: 2018 Journal: Acta Cryst. D / Year: 2018Title: Aminolipopeptide helioferin A and B Authors: Gessmann, R. / Petratos, K. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6evh.cif.gz 6evh.cif.gz | 18 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6evh.ent.gz pdb6evh.ent.gz | 11.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6evh.json.gz 6evh.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6evh_validation.pdf.gz 6evh_validation.pdf.gz | 402.7 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6evh_full_validation.pdf.gz 6evh_full_validation.pdf.gz | 402.7 KB | Display | |
| Data in XML |  6evh_validation.xml.gz 6evh_validation.xml.gz | 2.6 KB | Display | |
| Data in CIF |  6evh_validation.cif.gz 6evh_validation.cif.gz | 2.6 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ev/6evh https://data.pdbj.org/pub/pdb/validation_reports/ev/6evh ftp://data.pdbj.org/pub/pdb/validation_reports/ev/6evh ftp://data.pdbj.org/pub/pdb/validation_reports/ev/6evh | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
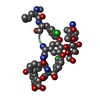
| Deposited unit | 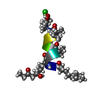
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein/peptide | Mass: 1121.496 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: MOO is 2-methyl-N-1-oxooctanoic acid,AMO is AHMOD is (2S, 4S, 6S)-2-amino-6-hydroxy-4-methyl-8-oxodecanoyl,AIB is alpha-aminoisobutyric acid,C9K is APAE is conformation A 2-(2-aminopropyl) ...Details: MOO is 2-methyl-N-1-oxooctanoic acid,AMO is AHMOD is (2S, 4S, 6S)-2-amino-6-hydroxy-4-methyl-8-oxodecanoyl,AIB is alpha-aminoisobutyric acid,C9K is APAE is conformation A 2-(2-aminopropyl)aminoethanol and C9N, conformation B AMAE is 2-[(2-aminopropyl)-methylamino]-ethanol Source: (natural)  Mycogone rosea (fungus) Mycogone rosea (fungus) |
|---|---|
| #2: Chemical | ChemComp-CL / |
| #3: Chemical | ChemComp-F / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.44 Å3/Da / Description: plate, 0.5 mm x 0.2 mm x 0.03 |
|---|---|
| Crystal grow | Temperature: 297 K / Method: evaporation / Details: slow evaporation in a closed vial |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source: SEALED TUBE / Type: BRUKER IMUS MICROFOCUS / Wavelength: 1.54178 Å |
| Detector | Type: RDI CMOS_8M / Detector: CMOS / Date: Jun 7, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.54178 Å / Relative weight: 1 |
| Reflection | Resolution: 0.82→17.3 Å / Num. obs: 11314 / % possible obs: 91.5 % / Redundancy: 9.7 % / Net I/σ(I): 26.2 |
| Reflection shell | Resolution: 0.9→1 Å |
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: AB INITIO PHASING / Resolution: 0.9→17.3 Å / Cross valid method: FREE R-VALUE Details: The last amino alcohol residue is Apae in helioferin A and Amae in helioferin B. The deposited structure is neutral The presence of a proton at N2 of the last residue Apae/Amae is implicated.
| ||||||||||||||||||||
| Displacement parameters | Biso mean: 8 Å2 | ||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 0.9→17.3 Å /
|
 Movie
Movie Controller
Controller






 PDBj
PDBj



