[English] 日本語
 Yorodumi
Yorodumi- PDB-6cro: CRYSTAL STRUCTURE OF LAMBDA-CRO BOUND TO A CONSENSUS OPERATOR AT ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6cro | ||||||
|---|---|---|---|---|---|---|---|
| Title | CRYSTAL STRUCTURE OF LAMBDA-CRO BOUND TO A CONSENSUS OPERATOR AT 3.0 ANGSTROM RESOLUTION | ||||||
 Components Components |
| ||||||
 Keywords Keywords | GENE REGULATION/DNA / COMPLEX (TRANSCRIPTION REGULATION-DNA) / CRO / BACTERIOPHAGE LAMBDA / CONFORMATIONAL CHANGE / REPRESSOR / HELIX-TURN-HELIX / GENE REGULATION-DNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationlatency-replication decision / release from viral latency / negative regulation of viral transcription / negative regulation of transcription by competitive promoter binding / core promoter sequence-specific DNA binding / response to UV / protein homodimerization activity / DNA binding Similarity search - Function | ||||||
| Biological species |  Enterobacteria phage lambda (virus) Enterobacteria phage lambda (virus) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MIR, WITH SUBSEQUENT MOLECULAR REPLACEMENT / Resolution: 3 Å MIR, WITH SUBSEQUENT MOLECULAR REPLACEMENT / Resolution: 3 Å | ||||||
 Authors Authors | Albright, R.A. / Matthews, B.W. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1998 Journal: J.Mol.Biol. / Year: 1998Title: Crystal structure of lambda-Cro bound to a consensus operator at 3.0 A resolution. Authors: Albright, R.A. / Matthews, B.W. #1:  Journal: Protein Sci. / Year: 1998 Journal: Protein Sci. / Year: 1998Title: Crystal Structure of an Engineered Cro Monomer Bound Nonspecifically to DNA: Possible Implications for Nonspecific Binding by the Wild-Type Protein Authors: Albright, R.A. / Mossing, M.C. / Matthews, B.W. #2:  Journal: J.Mol.Biol. / Year: 1998 Journal: J.Mol.Biol. / Year: 1998Title: Refined Structure of Cro Repressor Protein from Bacteriophage Lambda Suggests Both Flexibility and Plasticity Authors: Ohlendorf, D.H. / Tronrud, D.E. / Matthews, B.W. #3:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1998 Journal: Proc.Natl.Acad.Sci.USA / Year: 1998Title: How Cro and Lambda-Repressor Distinguish between Operators: The Structural Basis Underlying a Genetic Switch Authors: Albright, R.A. / Matthews, B.W. #4:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1990 Journal: Proc.Natl.Acad.Sci.USA / Year: 1990Title: Protein-DNA Conformational Changes in the Crystal Structure of a Lambda Cro- Operator Complex Authors: Brennan, R.G. / Roderick, S.L. / Takeda, Y. / Matthews, B.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6cro.cif.gz 6cro.cif.gz | 49.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6cro.ent.gz pdb6cro.ent.gz | 32.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6cro.json.gz 6cro.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6cro_validation.pdf.gz 6cro_validation.pdf.gz | 433.5 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6cro_full_validation.pdf.gz 6cro_full_validation.pdf.gz | 454.5 KB | Display | |
| Data in XML |  6cro_validation.xml.gz 6cro_validation.xml.gz | 8.7 KB | Display | |
| Data in CIF |  6cro_validation.cif.gz 6cro_validation.cif.gz | 11.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cr/6cro https://data.pdbj.org/pub/pdb/validation_reports/cr/6cro ftp://data.pdbj.org/pub/pdb/validation_reports/cr/6cro ftp://data.pdbj.org/pub/pdb/validation_reports/cr/6cro | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 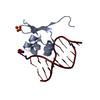
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 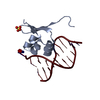
| ||||||||||
| Unit cell |
| ||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: DNA chain | Mass: 6118.968 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: DNA chain | Mass: 6149.979 Da / Num. of mol.: 1 / Source method: obtained synthetically |
| #3: Protein | Mass: 6712.643 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Enterobacteria phage lambda (virus) / Genus: Lambda-like viruses / Production host: Enterobacteria phage lambda (virus) / Genus: Lambda-like viruses / Production host:  |
| #4: Chemical | ChemComp-SO4 / |
| #5: Water | ChemComp-HOH / |
| Compound details | THIS ENTRY CONTAINS THE COMPLETE ASYMMETRIC UNIT. THE PROTEIN CHAIN IS PRESENTED AS CHAIN A WITH ...THIS ENTRY CONTAINS THE COMPLETE ASYMMETRIC |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.3 Å3/Da / Density % sol: 47.33 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 6.9 Details: CRO PROTEIN WAS SUSPENDED IN 20MM SODIUM CACODYLATE PH6.9 THEN MIXED WITH A 30% EXCESS OF THE 19BP DNA FRAGMENT. THE COMPLEX WAS THEN MIXED WITH AN EQUAL VOLUME OF PRECIPITANT SOLUTION (70MM ...Details: CRO PROTEIN WAS SUSPENDED IN 20MM SODIUM CACODYLATE PH6.9 THEN MIXED WITH A 30% EXCESS OF THE 19BP DNA FRAGMENT. THE COMPLEX WAS THEN MIXED WITH AN EQUAL VOLUME OF PRECIPITANT SOLUTION (70MM AMMONIUM SULFATE, 13% PEG3350) AND ALLOWED TO EQUILIBRATE VIA THE HANGING DROP METHOD AT ROOM TEMPERATURE. COCRYSTALS TYPICALLY TAKE 3-4 MONTHS TO APPEAR., vapor diffusion - hanging drop, temperature 293K | ||||||||||||||||||||||||||||||
| Components of the solutions |
| ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 295 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 ROTATING ANODE / Type: RIGAKU RU200 |
| Detector | Type: SDMS / Detector: AREA DETECTOR / Date: Feb 15, 1993 / Details: COLLIMATOR |
| Radiation | Monochromator: GRAPHITE CRYSTAL / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Relative weight: 1 |
| Reflection | Resolution: 3→20 Å / Num. all: 3758 / Num. obs: 3758 / % possible obs: 99.9 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 17.7 % / Biso Wilson estimate: 40.8 Å2 / Rsym value: 0.072 |
| Reflection shell | Highest resolution: 3 Å |
| Reflection | *PLUS Num. all: 3760 / Num. measured all: 132786 / Rmerge(I) obs: 0.072 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MIR, WITH SUBSEQUENT MOLECULAR REPLACEMENT MIR, WITH SUBSEQUENT MOLECULAR REPLACEMENTStarting model: TRUNCATED VERSION OF THE PREVIOUS LOW-RESOLUTION COMPLEX STRUCTURE Resolution: 3→20 Å / σ(F): 0 Stereochemistry target values: TNT PROTGEO & NUCLGEO ISOTROPIC THERMAL FACTOR RESTRAINTS: TNT BCORREL (MODIFIED TO INCLUDE DNA) Details: THIS MODEL UNDERWENT ONE FINAL ROUND OF REFINEMENT AFTER THE MANUSCRIPT WAS SUBMITTED. THE PSEUDO-DYAD OF THE COMPLEX IS COINCIDENT WITH A CRYSTALLOGRAPHIC TWO-FOLD AXIS, SUCH THAT THE ...Details: THIS MODEL UNDERWENT ONE FINAL ROUND OF REFINEMENT AFTER THE MANUSCRIPT WAS SUBMITTED. THE PSEUDO-DYAD OF THE COMPLEX IS COINCIDENT WITH A CRYSTALLOGRAPHIC TWO-FOLD AXIS, SUCH THAT THE COMPLEX IS EVENLY-DISTRIBUTED BETWEEN TWO ORIENTATIONS RELATED BY A 180-DEGREE ROTATION. THE SPACE GROUP IS I 21 3 WITH A SINGLE CRO SUBUNIT AND AN "AVERAGED" DNA HALF-FRAGMENT PER ASU. THIS AVERAGING AFFECTED ONLY THOSE BP IDENTITIES WHERE THE SEQUENCE PALINDROME BREAKS DOWN, WHICH OCCURS ONLY IN REGIONS NOT CONTACTED BY CRO. THE PROTEIN, DNA BACKBONE, AND THOSE BPS DIRECTLY CONTACTED BY CRO ARE EVERYWHERE "NORMAL". THE AVERAGED BASES CONSISTED OF THE TWO COMPONENT BASES AT A GIVEN POSITION, EACH BONDED TO THE SAME RIBOSE, BUT ALLOWED TO IGNORE THE PRESENCE OF EACH OTHER DURING REFINEMENT.
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: BABINET / Bsol: 160 Å2 / ksol: 0.5 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: TNT / Version: 5EB / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor all: 0.194 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller










 PDBj
PDBj









































