[English] 日本語
 Yorodumi
Yorodumi- PDB-6cc8: Crystal structure MBD3 MBD domain in complex with methylated CpG DNA -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6cc8 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure MBD3 MBD domain in complex with methylated CpG DNA | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | DNA BINDING PROTEIN/DNA / Structural Genomics Consortium / SGC / DNA BINDING PROTEIN-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationventricular cardiac muscle tissue development / NuRD complex / regulation of cell fate specification / regulation of stem cell differentiation / methyl-CpG binding / DNA methylation-dependent constitutive heterochromatin formation / RNA Polymerase I Transcription Initiation / embryonic organ development / Transcriptional regulation of brown and beige adipocyte differentiation by EBF2 / Regulation of TP53 Activity through Acetylation ...ventricular cardiac muscle tissue development / NuRD complex / regulation of cell fate specification / regulation of stem cell differentiation / methyl-CpG binding / DNA methylation-dependent constitutive heterochromatin formation / RNA Polymerase I Transcription Initiation / embryonic organ development / Transcriptional regulation of brown and beige adipocyte differentiation by EBF2 / Regulation of TP53 Activity through Acetylation / heterochromatin / Regulation of PTEN gene transcription / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / Regulation of endogenous retroelements by KRAB-ZFP proteins / HDACs deacetylate histones / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / response to nutrient levels / response to estradiol / Potential therapeutics for SARS / in utero embryonic development / chromatin remodeling / negative regulation of DNA-templated transcription / positive regulation of DNA-templated transcription / chromatin / negative regulation of transcription by RNA polymerase II / protein-containing complex / DNA binding / nucleoplasm / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human)synthetic construct (others) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.95 Å MOLECULAR REPLACEMENT / Resolution: 1.95 Å | |||||||||
 Authors Authors | Liu, K. / Tempel, W. / Bountra, C. / Arrowsmith, C.H. / Edwards, A.M. / Min, J. / Structural Genomics Consortium (SGC) | |||||||||
 Citation Citation |  Journal: Febs J. / Year: 2019 Journal: Febs J. / Year: 2019Title: Structural analyses reveal that MBD3 is a methylated CG binder. Authors: Liu, K. / Lei, M. / Wu, Z. / Gan, B. / Cheng, H. / Li, Y. / Min, J. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6cc8.cif.gz 6cc8.cif.gz | 124.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6cc8.ent.gz pdb6cc8.ent.gz | 93.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6cc8.json.gz 6cc8.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cc/6cc8 https://data.pdbj.org/pub/pdb/validation_reports/cc/6cc8 ftp://data.pdbj.org/pub/pdb/validation_reports/cc/6cc8 ftp://data.pdbj.org/pub/pdb/validation_reports/cc/6cc8 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6ccgC  6ceuC  6cevC  6c2k S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 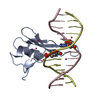
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 8473.716 Da / Num. of mol.: 2 / Fragment: MBD domain (UNP residues 1-71) Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: MBD3 / Plasmid: pET28-GSTLIC / Production host: Homo sapiens (human) / Gene: MBD3 / Plasmid: pET28-GSTLIC / Production host:  #2: DNA chain | Mass: 3677.419 Da / Num. of mol.: 4 / Source method: obtained synthetically / Details: dodecanucleotide / Source: (synth.) synthetic construct (others) #3: Chemical | ChemComp-UNX / #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.8 Å3/Da / Density % sol: 55.4 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion / pH: 7.5 Details: 25% PEG3350, 0.2 M sodium chloride, 0.1 M HEPES, 5% ethylene glycol |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-ID / Wavelength: 0.97923 Å / Beamline: 19-ID / Wavelength: 0.97923 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Oct 20, 2012 |
| Radiation | Monochromator: double crystal Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97923 Å / Relative weight: 1 |
| Reflection | Resolution: 1.95→35.73 Å / Num. obs: 25160 / % possible obs: 99.9 % / Redundancy: 5.3 % / Rmerge(I) obs: 0.074 / Net I/σ(I): 14 |
| Reflection shell | Resolution: 1.95→2 Å / Redundancy: 3.7 % / Rmerge(I) obs: 0.761 / % possible all: 99.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 6C2K  6c2k Resolution: 1.95→35.73 Å / Cor.coef. Fo:Fc: 0.949 / Cor.coef. Fo:Fc free: 0.935 / SU B: 9.689 / SU ML: 0.132 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.17 / ESU R Free: 0.156 Details: MODEL WAS ORIGINALLY REFINED AGAINST DATA COLLECTED ON AN ISOMORPHOUS CRYSTAL. THAT STRUCTURE WAS SOLVED BY MOLECULAR REPLACEMENT USING PHASER AND MOLREP TO POSITION FIRST AND SECOND COPY OF ...Details: MODEL WAS ORIGINALLY REFINED AGAINST DATA COLLECTED ON AN ISOMORPHOUS CRYSTAL. THAT STRUCTURE WAS SOLVED BY MOLECULAR REPLACEMENT USING PHASER AND MOLREP TO POSITION FIRST AND SECOND COPY OF THE COMPLEX, RESPECTIVELY. REFMAC WAS USED DURING INTERMEDIATE REFINEMENT. COOT WAS USED FOR INTERACTIVE MODEL BUILDING. MODEL GEOMETRY WAS ASSESSED ON THE MOLPROBITY SERVER.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 42.99 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.95→35.73 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj











































