+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6b8o | ||||||
|---|---|---|---|---|---|---|---|
| Title | WT Ig-like V Domain with Phosphatidylserine | ||||||
 Components Components | Triggering receptor expressed on myeloid cells 2 | ||||||
 Keywords Keywords | SIGNALING PROTEIN / Membrane receptor | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of high-density lipoprotein particle clearance / regulation of toll-like receptor 6 signaling pathway / positive regulation of complement activation, classical pathway / detection of lipoteichoic acid / regulation of macrophage inflammatory protein 1 alpha production / regulation of hippocampal neuron apoptotic process / regulation of plasma membrane bounded cell projection organization / positive regulation of C-C chemokine receptor CCR7 signaling pathway / excitatory synapse pruning / positive regulation of CD40 signaling pathway ...positive regulation of high-density lipoprotein particle clearance / regulation of toll-like receptor 6 signaling pathway / positive regulation of complement activation, classical pathway / detection of lipoteichoic acid / regulation of macrophage inflammatory protein 1 alpha production / regulation of hippocampal neuron apoptotic process / regulation of plasma membrane bounded cell projection organization / positive regulation of C-C chemokine receptor CCR7 signaling pathway / excitatory synapse pruning / positive regulation of CD40 signaling pathway / negative regulation of triglyceride storage / negative regulation of cell activation / detection of peptidoglycan / positive regulation of macrophage fusion / import into cell / sulfatide binding / positive regulation of establishment of protein localization / negative regulation of fat cell proliferation / lipoteichoic acid binding / positive regulation of engulfment of apoptotic cell / negative regulation of macrophage colony-stimulating factor signaling pathway / positive regulation of synapse pruning / microglial cell activation involved in immune response / positive regulation of antigen processing and presentation of peptide antigen via MHC class II / negative regulation of toll-like receptor 2 signaling pathway / negative regulation of autophagic cell death / respiratory burst after phagocytosis / negative regulation of astrocyte activation / positive regulation of CAMKK-AMPK signaling cascade / semaphorin receptor binding / positive regulation of low-density lipoprotein particle clearance / positive regulation of microglial cell migration / detection of lipopolysaccharide / Other semaphorin interactions / T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell / apolipoprotein A-I binding / CXCL12-activated CXCR4 signaling pathway / negative regulation of toll-like receptor 4 signaling pathway / negative regulation of neuroinflammatory response / negative regulation of p38MAPK cascade / very-low-density lipoprotein particle binding / high-density lipoprotein particle binding / cellular response to oxidised low-density lipoprotein particle stimulus / negative regulation of glial cell apoptotic process / dendritic cell differentiation / microglial cell proliferation / complement-mediated synapse pruning / positive regulation of microglial cell activation / phagocytosis, recognition / amyloid-beta clearance by cellular catabolic process / semaphorin receptor complex / low-density lipoprotein particle binding / cellular response to lipoprotein particle stimulus / negative regulation of NLRP3 inflammasome complex assembly / regulation of resting membrane potential / cellular response to peptidoglycan / positive regulation of phagocytosis, engulfment / peptidoglycan binding / regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway / positive regulation of amyloid-beta clearance / positive regulation of chemotaxis / regulation of TOR signaling / semaphorin receptor activity / positive regulation of potassium ion transport / phosphatidylethanolamine binding / positive regulation of proteasomal protein catabolic process / positive regulation of osteoclast differentiation / negative regulation of amyloid fibril formation / cellular response to lipid / regulation of interleukin-6 production / kinase activator activity / dendritic spine maintenance / apoptotic cell clearance / negative regulation of interleukin-1 beta production / phagocytosis, engulfment / phosphatidylserine binding / regulation of innate immune response / regulation of cytokine production involved in inflammatory response / negative regulation of cholesterol storage / cellular response to lipoteichoic acid / lipid homeostasis / lipoprotein particle binding / amyloid-beta clearance / positive regulation of ATP biosynthetic process / humoral immune response / pyroptotic inflammatory response / positive regulation of intracellular signal transduction / regulation of lipid metabolic process / positive regulation of interleukin-10 production / apolipoprotein binding / negative regulation of tumor necrosis factor production / plasma membrane raft / positive regulation of cholesterol efflux / social behavior / positive regulation of TOR signaling / response to axon injury / negative regulation of cytokine production involved in inflammatory response / astrocyte activation / positive regulation of calcium-mediated signaling / negative regulation of autophagy Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å | ||||||
 Authors Authors | Sudom, A. / Wang, Z. | ||||||
 Citation Citation |  Journal: J. Biol. Chem. / Year: 2018 Journal: J. Biol. Chem. / Year: 2018Title: Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2. Authors: Sudom, A. / Talreja, S. / Danao, J. / Bragg, E. / Kegel, R. / Min, X. / Richardson, J. / Zhang, Z. / Sharkov, N. / Marcora, E. / Thibault, S. / Bradley, J. / Wood, S. / Lim, A.C. / Chen, H. ...Authors: Sudom, A. / Talreja, S. / Danao, J. / Bragg, E. / Kegel, R. / Min, X. / Richardson, J. / Zhang, Z. / Sharkov, N. / Marcora, E. / Thibault, S. / Bradley, J. / Wood, S. / Lim, A.C. / Chen, H. / Wang, S. / Foltz, I.N. / Sambashivan, S. / Wang, Z. #1:  Journal: To Be Published Journal: To Be PublishedTitle: Structure of Ig-like V domain in complex with phosphatidylserine to 2.2 Angstrom resolution Authors: Sudom, A. / Wang, Z. / Min, X. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6b8o.cif.gz 6b8o.cif.gz | 152.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6b8o.ent.gz pdb6b8o.ent.gz | 117.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6b8o.json.gz 6b8o.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b8/6b8o https://data.pdbj.org/pub/pdb/validation_reports/b8/6b8o ftp://data.pdbj.org/pub/pdb/validation_reports/b8/6b8o ftp://data.pdbj.org/pub/pdb/validation_reports/b8/6b8o | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5ud7SC  5ud8C S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein / Sugars , 2 types, 12 molecules ABCDEF

| #1: Protein | Mass: 19138.254 Da / Num. of mol.: 6 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TREM2 / Production host: Homo sapiens (human) / Gene: TREM2 / Production host:  Homo sapiens (human) / References: UniProt: Q9NZC2 Homo sapiens (human) / References: UniProt: Q9NZC2#2: Sugar | ChemComp-NAG / |
|---|
-Non-polymers , 5 types, 113 molecules 


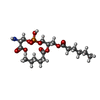





| #3: Chemical | ChemComp-IOD / #4: Chemical | ChemComp-SO4 / #5: Chemical | ChemComp-EDO / #6: Chemical | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.41 Å3/Da / Density % sol: 49.01 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion / Details: 0.2 M NaI, 1.6 M ammonium sulfate, 50 mM Tris 8.5 |
-Data collection
| Diffraction | Mean temperature: 80 K | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 5.0.2 / Wavelength: 1.00001 Å / Beamline: 5.0.2 / Wavelength: 1.00001 Å | ||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: Jul 28, 2017 | ||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.00001 Å / Relative weight: 1 | ||||||||||||||||||||||||
| Reflection | Resolution: 2.2→47.36 Å / Num. obs: 57469 / % possible obs: 100 % / Redundancy: 13.1 % / CC1/2: 0.999 / Rmerge(I) obs: 0.106 / Net I/σ(I): 19.8 / Num. measured all: 752391 / Scaling rejects: 0 | ||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5UD7 Resolution: 2.2→47.36 Å / Cor.coef. Fo:Fc: 0.94 / Cor.coef. Fo:Fc free: 0.918 / SU B: 5.77 / SU ML: 0.142 / SU R Cruickshank DPI: 0.201 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.201 / ESU R Free: 0.184 Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : REFINED INDIVIDUALLY
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 148.23 Å2 / Biso mean: 52.028 Å2 / Biso min: 19.21 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.2→47.36 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.2→2.257 Å / Rfactor Rfree error: 0 / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller











 PDBj
PDBj





