Entry Database : PDB / ID : 5tupTitle X-ray Crystal Structure of the Aspergillus fumigatus Sliding Clamp Proliferating cell nuclear antigen Keywords / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Aspergillus fumigatus Z5 (mold)Method / / / / Resolution : 2.602 Å Authors Bruning, J.B. / Marshall, A.C. / Kroker, A.J. / Wegener, K.L. / Rajapaksha, H. Journal : FEBS J. / Year : 2017Title : Structure of the sliding clamp from the fungal pathogen Aspergillus fumigatus (AfumPCNA) and interactions with Human p21.Authors : Marshall, A.C. / Kroker, A.J. / Murray, L.A. / Gronthos, K. / Rajapaksha, H. / Wegener, K.L. / Bruning, J.B. History Deposition Nov 6, 2016 Deposition site / Processing site Revision 1.0 Feb 22, 2017 Provider / Type Revision 1.1 Mar 29, 2017 Group Revision 1.2 Nov 1, 2017 Group / Data collectionCategory / pdbx_struct_assembly_auth_evidenceItem Revision 1.3 Oct 4, 2023 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_ncs_dom_lim Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.602 Å
molecular replacement / Resolution: 2.602 Å  Authors
Authors Citation
Citation Journal: FEBS J. / Year: 2017
Journal: FEBS J. / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5tup.cif.gz
5tup.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5tup.ent.gz
pdb5tup.ent.gz PDB format
PDB format 5tup.json.gz
5tup.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/tu/5tup
https://data.pdbj.org/pub/pdb/validation_reports/tu/5tup ftp://data.pdbj.org/pub/pdb/validation_reports/tu/5tup
ftp://data.pdbj.org/pub/pdb/validation_reports/tu/5tup
 Links
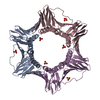
Links Assembly
Assembly
 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Australian Synchrotron
Australian Synchrotron  / Beamline: MX1 / Wavelength: 0.9537 Å
/ Beamline: MX1 / Wavelength: 0.9537 Å molecular replacement
molecular replacement Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj

