+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5ta3 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 2) | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSPORT PROTEIN/ISOMERASE / RyR / Ca2+ / EC coupling / gating / TRANSPORT PROTEIN-ISOMERASE complex | ||||||
| Function / homology |  Function and homology information Function and homology informationATP-gated ion channel activity / : / negative regulation of calcium-mediated signaling / negative regulation of insulin secretion involved in cellular response to glucose stimulus / neuronal action potential propagation / negative regulation of release of sequestered calcium ion into cytosol / insulin secretion involved in cellular response to glucose stimulus / terminal cisterna / ryanodine-sensitive calcium-release channel activity / ryanodine receptor complex ...ATP-gated ion channel activity / : / negative regulation of calcium-mediated signaling / negative regulation of insulin secretion involved in cellular response to glucose stimulus / neuronal action potential propagation / negative regulation of release of sequestered calcium ion into cytosol / insulin secretion involved in cellular response to glucose stimulus / terminal cisterna / ryanodine-sensitive calcium-release channel activity / ryanodine receptor complex / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / response to redox state / ossification involved in bone maturation / negative regulation of heart rate / cellular response to caffeine / 'de novo' protein folding / skin development / FK506 binding / organelle membrane / intracellularly gated calcium channel activity / smooth endoplasmic reticulum / outflow tract morphogenesis / smooth muscle contraction / T cell proliferation / toxic substance binding / striated muscle contraction / regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion / voltage-gated calcium channel activity / calcium channel inhibitor activity / skeletal muscle fiber development / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / Ion homeostasis / release of sequestered calcium ion into cytosol / calcium channel complex / sarcoplasmic reticulum membrane / cellular response to calcium ion / muscle contraction / sarcoplasmic reticulum / protein maturation / peptidylprolyl isomerase / calcium channel regulator activity / peptidyl-prolyl cis-trans isomerase activity / calcium-mediated signaling / sarcolemma / calcium ion transmembrane transport / Stimuli-sensing channels / calcium channel activity / Z disc / intracellular calcium ion homeostasis / disordered domain specific binding / positive regulation of cytosolic calcium ion concentration / protein refolding / protein homotetramerization / transmembrane transporter binding / calmodulin binding / signaling receptor binding / calcium ion binding / ATP binding / identical protein binding / membrane / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 4.4 Å | ||||||
 Authors Authors | Clarke, O.B. / des Georges, A. / Zalk, R. / Marks, A.R. / Hendrickson, W.A. / Frank, J. | ||||||
 Citation Citation |  Journal: Cell / Year: 2016 Journal: Cell / Year: 2016Title: Structural Basis for Gating and Activation of RyR1. Authors: Amédée des Georges / Oliver B Clarke / Ran Zalk / Qi Yuan / Kendall J Condon / Robert A Grassucci / Wayne A Hendrickson / Andrew R Marks / Joachim Frank /  Abstract: The type-1 ryanodine receptor (RyR1) is an intracellular calcium (Ca(2+)) release channel required for skeletal muscle contraction. Here, we present cryo-EM reconstructions of RyR1 in multiple ...The type-1 ryanodine receptor (RyR1) is an intracellular calcium (Ca(2+)) release channel required for skeletal muscle contraction. Here, we present cryo-EM reconstructions of RyR1 in multiple functional states revealing the structural basis of channel gating and ligand-dependent activation. Binding sites for the channel activators Ca(2+), ATP, and caffeine were identified at interdomain interfaces of the C-terminal domain. Either ATP or Ca(2+) alone induces conformational changes in the cytoplasmic assembly ("priming"), without pore dilation. In contrast, in the presence of all three activating ligands, high-resolution reconstructions of open and closed states of RyR1 were obtained from the same sample, enabling analyses of conformational changes associated with gating. Gating involves global conformational changes in the cytosolic assembly accompanied by local changes in the transmembrane domain, which include bending of the S6 transmembrane segment and consequent pore dilation, displacement, and deformation of the S4-S5 linker and conformational changes in the pseudo-voltage-sensor domain. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5ta3.cif.gz 5ta3.cif.gz | 2.7 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5ta3.ent.gz pdb5ta3.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  5ta3.json.gz 5ta3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ta/5ta3 https://data.pdbj.org/pub/pdb/validation_reports/ta/5ta3 ftp://data.pdbj.org/pub/pdb/validation_reports/ta/5ta3 ftp://data.pdbj.org/pub/pdb/validation_reports/ta/5ta3 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  8377MC  8342C  8372C  8373C  8374C  8375C  8376C  8378C  8379C  8380C  8381C  8382C  8383C  8384C  8385C  8386C  8387C  8388C  8389C  8390C  8391C  8392C  8393C  8394C  8395C  5t15C  5t9mC  5t9nC  5t9rC  5t9sC  5t9vC  5talC  5tamC  5tanC  5tapC  5taqC  5tasC  5tatC  5tauC  5tavC  5tawC  5taxC  5tayC  5tazC  5tb0C  5tb1C  5tb2C  5tb3C  5tb4C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 2 types, 8 molecules FAHJBGIE
| #1: Protein | Mass: 11798.501 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: FKBP1B, FKBP12.6, FKBP1L, FKBP9, OTK4 / Production host: Homo sapiens (human) / Gene: FKBP1B, FKBP12.6, FKBP1L, FKBP9, OTK4 / Production host:  #2: Protein | Mass: 475107.719 Da / Num. of mol.: 4 / Source method: isolated from a natural source / Source: (natural)  |
|---|
-Non-polymers , 4 types, 16 molecules 
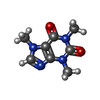





| #3: Chemical | ChemComp-ATP / #4: Chemical | ChemComp-CFF / #5: Chemical | ChemComp-ZN / #6: Chemical | ChemComp-CA / |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: RyR1-Cs2 complex / Type: COMPLEX / Entity ID: #1-#2 / Source: MULTIPLE SOURCES |
|---|---|
| Buffer solution | pH: 7.4 |
| Specimen | Conc.: 6 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Grid material: GOLD / Grid mesh size: 400 divisions/in. / Grid type: Quantifoil |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K Details: Blotted for 3-4 seconds on both sides with Whatman ashless filter paper, blot force 3, wait time 30 seconds |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI POLARA 300 |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Image recording | Electron dose: 50 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 4.4 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 55564 / Algorithm: FOURIER SPACE Details: The reported resolution is for the core. The resolution of the whole assembly is 4.6 Angstrom. Symmetry type: POINT |
 Movie
Movie Controller
Controller













 PDBj
PDBj








