Entry Database : PDB / ID : 5t5tTitle AMPK bound to allosteric activator (5'-AMP-activated protein kinase subunit ...) x 2 5'-AMP-activated protein kinase catalytic subunit alpha-1 Keywords / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Rattus norvegicus (Norway rat)Method / / / / Resolution : 3.46 Å Authors Calabrese, M.F. / Kurumbail, R.G. Journal : J. Pharmacol. Exp. Ther. / Year : 2017Title : Selective Activation of AMPK beta 1-Containing Isoforms Improves Kidney Function in a Rat Model of Diabetic Nephropathy.Authors: Salatto, C.T. / Miller, R.A. / Cameron, K.O. / Cokorinos, E. / Reyes, A. / Ward, J. / Calabrese, M.F. / Kurumbail, R.G. / Rajamohan, F. / Kalgutkar, A.S. / Tess, D.A. / Shavnya, A. / Genung, ... Authors : Salatto, C.T. / Miller, R.A. / Cameron, K.O. / Cokorinos, E. / Reyes, A. / Ward, J. / Calabrese, M.F. / Kurumbail, R.G. / Rajamohan, F. / Kalgutkar, A.S. / Tess, D.A. / Shavnya, A. / Genung, N.E. / Edmonds, D.J. / Jatkar, A. / Maciejewski, B.S. / Amaro, M. / Gandhok, H. / Monetti, M. / Cialdea, K. / Bollinger, E. / Kreeger, J.M. / Coskran, T.M. / Opsahl, A.C. / Boucher, G.G. / Birnbaum, M.J. / DaSilva-Jardine, P. / Rolph, T. History Deposition Aug 31, 2016 Deposition site / Processing site Revision 1.0 Mar 29, 2017 Provider / Type Revision 1.1 Apr 26, 2017 Group Revision 1.2 Oct 4, 2023 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_ref_seq_dif Item / _database_2.pdbx_database_accession / _struct_ref_seq_dif.detailsRevision 1.3 Nov 6, 2024 Group / Category / pdbx_modification_feature
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 3.46 Å
molecular replacement / Resolution: 3.46 Å  Authors
Authors Citation
Citation Journal: J. Pharmacol. Exp. Ther. / Year: 2017
Journal: J. Pharmacol. Exp. Ther. / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5t5t.cif.gz
5t5t.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5t5t.ent.gz
pdb5t5t.ent.gz PDB format
PDB format 5t5t.json.gz
5t5t.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/t5/5t5t
https://data.pdbj.org/pub/pdb/validation_reports/t5/5t5t ftp://data.pdbj.org/pub/pdb/validation_reports/t5/5t5t
ftp://data.pdbj.org/pub/pdb/validation_reports/t5/5t5t
 Links
Links Assembly
Assembly

 Components
Components







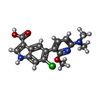







 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  APS
APS  / Beamline: 17-ID / Wavelength: 1 Å
/ Beamline: 17-ID / Wavelength: 1 Å molecular replacement
molecular replacement Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller













 PDBj
PDBj











