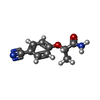
Entry Database : PDB / ID : 5rlgTitle PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z19739650 Helicase Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Method / / / / Resolution : 1.96 Å Authors Newman, J.A. / Yosaatmadja, Y. / Douangamath, A. / Aimon, A. / Powell, A.J. / Dias, A. / Fearon, D. / Dunnett, L. / Brandao-Neto, J. / Krojer, T. ...Newman, J.A. / Yosaatmadja, Y. / Douangamath, A. / Aimon, A. / Powell, A.J. / Dias, A. / Fearon, D. / Dunnett, L. / Brandao-Neto, J. / Krojer, T. / Skyner, R. / Gorrie-Stone, T. / Thompson, W. / von Delft, F. / Arrowsmith, C.H. / Edwards, A. / Bountra, C. / Gileadi, O. Journal : Nat Commun / Year : 2021Title : Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.Authors : Newman, J.A. / Douangamath, A. / Yadzani, S. / Yosaatmadja, Y. / Aimon, A. / Brandao-Neto, J. / Dunnett, L. / Gorrie-Stone, T. / Skyner, R. / Fearon, D. / Schapira, M. / von Delft, F. / Gileadi, O. History Deposition Sep 16, 2020 Deposition site / Processing site Revision 1.0 Sep 30, 2020 Provider / Type Revision 1.1 Sep 7, 2022 Group / Category / citation_author / database_2Item _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year / _database_2.pdbx_DOI / _database_2.pdbx_database_accession Revision 1.2 May 22, 2024 Group / Category / chem_comp_bondRevision 1.3 Feb 18, 2026 Group / Structure summaryCategory / pdbx_initial_refinement_modelItem
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  FOURIER SYNTHESIS /
FOURIER SYNTHESIS /  molecular replacement / Resolution: 1.96 Å
molecular replacement / Resolution: 1.96 Å  Authors
Authors Citation
Citation Journal: Nat Commun / Year: 2021
Journal: Nat Commun / Year: 2021 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5rlg.cif.gz
5rlg.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5rlg.ent.gz
pdb5rlg.ent.gz PDB format
PDB format 5rlg.json.gz
5rlg.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/rl/5rlg
https://data.pdbj.org/pub/pdb/validation_reports/rl/5rlg ftp://data.pdbj.org/pub/pdb/validation_reports/rl/5rlg
ftp://data.pdbj.org/pub/pdb/validation_reports/rl/5rlg
 Links
Links Assembly
Assembly


 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I04-1 / Wavelength: 0.9126 Å
/ Beamline: I04-1 / Wavelength: 0.9126 Å molecular replacement
molecular replacement Processing
Processing FOURIER SYNTHESIS
FOURIER SYNTHESIS Movie
Movie Controller
Controller































 PDBj
PDBj