+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5qao | ||||||
|---|---|---|---|---|---|---|---|
| Title | OXA-48 IN COMPLEX WITH COMPOUND 19b | ||||||
 Components Components | Beta-lactamase | ||||||
 Keywords Keywords | HYDROLASE / Oxacillinase / Inhibitor / Complex / OXA / Antibiotic resistance / beta-lactamase / fragment | ||||||
| Function / homology |  Function and homology information Function and homology informationpenicillin binding / antibiotic catabolic process / cell wall organization / beta-lactamase activity / beta-lactamase / response to antibiotic / plasma membrane Similarity search - Function | ||||||
| Biological species |  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Lund, B.A. / Leiros, H.K.S. | ||||||
 Citation Citation |  Journal: Eur J Med Chem / Year: 2018 Journal: Eur J Med Chem / Year: 2018Title: A focused fragment library targeting the antibiotic resistance enzyme - Oxacillinase-48: Synthesis, structural evaluation and inhibitor design. Authors: Akhter, S. / Lund, B.A. / Ismael, A. / Langer, M. / Isaksson, J. / Christopeit, T. / Leiros, H.S. / Bayer, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5qao.cif.gz 5qao.cif.gz | 585.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5qao.ent.gz pdb5qao.ent.gz | 494.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5qao.json.gz 5qao.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qa/5qao https://data.pdbj.org/pub/pdb/validation_reports/qa/5qao ftp://data.pdbj.org/pub/pdb/validation_reports/qa/5qao ftp://data.pdbj.org/pub/pdb/validation_reports/qa/5qao | HTTPS FTP |
|---|
-Group deposition
| ID | G_1002027 (37 entries) |
|---|---|
| Title | A focused fragment library targeting the antibiotic resistance enzyme - oxacillinase-48: synthesis, structural evaluation and inhibitor design |
| Type | undefined |
| Description | A focused fragment library targeting the antibiotic resistance enzyme - oxacillinase-48: synthesis, structural evaluation and inhibitor design |
-Related structure data
| Related structure data |  5dtkS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 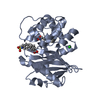
| |||||||||||||||||||||||||||||||||||
| 2 | 
| |||||||||||||||||||||||||||||||||||
| 3 | 
| |||||||||||||||||||||||||||||||||||
| 4 | 
| |||||||||||||||||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: 1 / Ens-ID: 1 / Beg auth comp-ID: GLU / Beg label comp-ID: GLU / End auth comp-ID: PRO / End label comp-ID: PRO / Auth seq-ID: 24 - 265 / Label seq-ID: 2 - 243
|
- Components
Components
-Protein , 1 types, 4 molecules ABCD
| #1: Protein | Mass: 28226.951 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Klebsiella pneumoniae (bacteria) / Gene: bla OXA-48, blaOXA-48, KPE71T_00045 / Production host: Klebsiella pneumoniae (bacteria) / Gene: bla OXA-48, blaOXA-48, KPE71T_00045 / Production host:  |
|---|
-Non-polymers , 5 types, 745 molecules 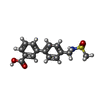








| #2: Chemical | ChemComp-Z8R / #3: Chemical | #4: Chemical | ChemComp-DMS / | #5: Chemical | #6: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.64 Å3/Da / Density % sol: 53.45 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 7.5 Details: 0.1 M HEPES pH 7.5, 8-11% PEG 8000 and 4-8% 1-butanol |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID23-1 / Wavelength: 0.98401 Å / Beamline: ID23-1 / Wavelength: 0.98401 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Sep 19, 2016 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.98401 Å / Relative weight: 1 |
| Reflection | Resolution: 2→62.66 Å / % possible obs: 96.78 % / Redundancy: 4.3 % / Rmerge(I) obs: 0.1512 / Rrim(I) all: 0.17 / Net I/σ(I): 3.8 |
| Reflection shell | Resolution: 2→2.07 Å / % possible obs: 98.7 % / Redundancy: 4.3 % / Rmerge(I) obs: 0.5944 / Num. unique obs: 34116 / Rrim(I) all: 0.67 / Net I/σ(I) obs: 1.64 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY: 5dtk Resolution: 2→62.66 Å / Cross valid method: THROUGHOUT
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2→62.66 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 28
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller
































 PDBj
PDBj



