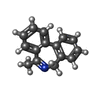
Entry Database : PDB / ID : 5ojlTitle Imine Reductase from Aspergillus terreus in complex with NADPH4 and dibenz[c,e]azepine Imine reductase Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Aspergillus terreus (mold)Method / / / Resolution : 1.56 Å Authors Sharma, M. / Grogan, G. Funding support Organization Grant number Country Biotechnology and Biological Sciences Research Council BB/M006832/1
Journal : Angew. Chem. Int. Ed. Engl. / Year : 2017Title : Biocatalytic Routes to Enantiomerically Enriched Dibenz[c,e]azepines.Authors : France, S.P. / Aleku, G.A. / Sharma, M. / Mangas-Sanchez, J. / Howard, R.M. / Steflik, J. / Kumar, R. / Adams, R.W. / Slabu, I. / Crook, R. / Grogan, G. / Wallace, T.W. / Turner, N.J. History Deposition Jul 21, 2017 Deposition site / Processing site Revision 1.0 May 30, 2018 Provider / Type Revision 1.1 Jan 17, 2024 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model Item / _database_2.pdbx_database_accession
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.56 Å
MOLECULAR REPLACEMENT / Resolution: 1.56 Å  Authors
Authors United Kingdom, 1items
United Kingdom, 1items  Citation
Citation Journal: Angew. Chem. Int. Ed. Engl. / Year: 2017
Journal: Angew. Chem. Int. Ed. Engl. / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5ojl.cif.gz
5ojl.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5ojl.ent.gz
pdb5ojl.ent.gz PDB format
PDB format 5ojl.json.gz
5ojl.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/oj/5ojl
https://data.pdbj.org/pub/pdb/validation_reports/oj/5ojl ftp://data.pdbj.org/pub/pdb/validation_reports/oj/5ojl
ftp://data.pdbj.org/pub/pdb/validation_reports/oj/5ojl
 Links
Links Assembly
Assembly

 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I04-1 / Wavelength: 0.92819 Å
/ Beamline: I04-1 / Wavelength: 0.92819 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj