| Entry | Database: PDB / ID: 5lhf
|
|---|
| Title | Phosphoribosyl anthranilate isomerase from Thermococcus kodakaraensis |
|---|
 Components Components | N-(5'-phosphoribosyl)anthranilate isomerase |
|---|
 Keywords Keywords | ISOMERASE / tryptophan biosynthesis / TIM barrel / protein stability |
|---|
| Function / homology |  Function and homology information Function and homology information
phosphoribosylanthranilate isomerase / phosphoribosylanthranilate isomerase activity / L-tryptophan biosynthetic processSimilarity search - Function N-(5'-phosphoribosyl)anthranilate isomerase family / N-(5'phosphoribosyl) anthranilate isomerase (PRAI) domain / N-(5'phosphoribosyl)anthranilate (PRA) isomerase / Ribulose-phosphate binding barrel / Aldolase class I / Aldolase-type TIM barrel / TIM Barrel / Alpha-Beta Barrel / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |   Thermococcus kodakarensis (archaea) Thermococcus kodakarensis (archaea) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.75 Å molecular replacement / Resolution: 1.75 Å |
|---|
 Authors Authors | Perveen, S. / Rashid, N. / Papageorgiou, A.C. |
|---|
 Citation Citation |  Journal: Acta Crystallogr F Struct Biol Commun / Year: 2016 Journal: Acta Crystallogr F Struct Biol Commun / Year: 2016
Title: Crystal structure of a phosphoribosyl anthranilate isomerase from the hyperthermophilic archaeon Thermococcus kodakaraensis.
Authors: Perveen, S. / Rashid, N. / Papageorgiou, A.C. |
|---|
| History | | Deposition | Jul 11, 2016 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Nov 9, 2016 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Dec 21, 2016 | Group: Database references |
|---|
| Revision 1.2 | Jan 10, 2024 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_symmetry / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_symmetry / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr1_symmetry / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn.ptnr2_symmetry |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 Thermococcus kodakarensis (archaea)
Thermococcus kodakarensis (archaea) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.75 Å
molecular replacement / Resolution: 1.75 Å  Authors
Authors Citation
Citation Journal: Acta Crystallogr F Struct Biol Commun / Year: 2016
Journal: Acta Crystallogr F Struct Biol Commun / Year: 2016 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5lhf.cif.gz
5lhf.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5lhf.ent.gz
pdb5lhf.ent.gz PDB format
PDB format 5lhf.json.gz
5lhf.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/lh/5lhf
https://data.pdbj.org/pub/pdb/validation_reports/lh/5lhf ftp://data.pdbj.org/pub/pdb/validation_reports/lh/5lhf
ftp://data.pdbj.org/pub/pdb/validation_reports/lh/5lhf
 Links
Links Assembly
Assembly
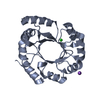

 Components
Components
 Thermococcus kodakarensis (strain ATCC BAA-918 / JCM 12380 / KOD1) (archaea)
Thermococcus kodakarensis (strain ATCC BAA-918 / JCM 12380 / KOD1) (archaea)
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ESRF
ESRF  / Beamline: MASSIF-1 / Wavelength: 0.96598 Å
/ Beamline: MASSIF-1 / Wavelength: 0.96598 Å molecular replacement
molecular replacement Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj




