+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5k7g | ||||||
|---|---|---|---|---|---|---|---|
| Title | IRAK4 in complex with AZ3862 | ||||||
 Components Components | Interleukin-1 receptor-associated kinase 4 | ||||||
 Keywords Keywords | TRANSFERASE/TRANSFERASE INHIBITOR / Inhibitor / Kinase / Protein tyrosine kinase / TRANSFERASE-TRANSFERASE INHIBITOR complex | ||||||
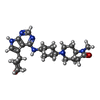
| Function / homology |  Function and homology information Function and homology informationIRAK4 deficiency (TLR5) / MyD88 dependent cascade initiated on endosome / TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation / MyD88 cascade initiated on plasma membrane / Toll signaling pathway / interleukin-33-mediated signaling pathway / neutrophil migration / toll-like receptor 9 signaling pathway / neutrophil mediated immunity / interleukin-1 receptor binding ...IRAK4 deficiency (TLR5) / MyD88 dependent cascade initiated on endosome / TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation / MyD88 cascade initiated on plasma membrane / Toll signaling pathway / interleukin-33-mediated signaling pathway / neutrophil migration / toll-like receptor 9 signaling pathway / neutrophil mediated immunity / interleukin-1 receptor binding / interleukin-1-mediated signaling pathway / extrinsic component of plasma membrane / IRAK4 deficiency (TLR2/4) / MyD88-dependent toll-like receptor signaling pathway / MyD88:MAL(TIRAP) cascade initiated on plasma membrane / toll-like receptor 4 signaling pathway / toll-like receptor signaling pathway / JNK cascade / TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling / lipopolysaccharide-mediated signaling pathway / positive regulation of smooth muscle cell proliferation / cytokine-mediated signaling pathway / Interleukin-1 signaling / kinase activity / PIP3 activates AKT signaling / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / eukaryotic translation initiation factor 2alpha kinase activity / positive regulation of canonical NF-kappaB signal transduction / 3-phosphoinositide-dependent protein kinase activity / DNA-dependent protein kinase activity / ribosomal protein S6 kinase activity / histone H3S10 kinase activity / histone H2AXS139 kinase activity / histone H3S28 kinase activity / histone H4S1 kinase activity / histone H2BS14 kinase activity / histone H3T3 kinase activity / histone H2AS121 kinase activity / Rho-dependent protein serine/threonine kinase activity / histone H2BS36 kinase activity / histone H3S57 kinase activity / histone H2AT120 kinase activity / AMP-activated protein kinase activity / histone H2AS1 kinase activity / histone H3T6 kinase activity / histone H3T11 kinase activity / histone H3T45 kinase activity / non-specific serine/threonine protein kinase / endosome membrane / intracellular signal transduction / innate immune response / protein serine kinase activity / protein serine/threonine kinase activity / protein kinase binding / cell surface / magnesium ion binding / extracellular space / ATP binding / nucleus / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.23 Å MOLECULAR REPLACEMENT / Resolution: 2.23 Å | ||||||
 Authors Authors | Ferguson, A.D. | ||||||
 Citation Citation |  Journal: J. Med. Chem. / Year: 2017 Journal: J. Med. Chem. / Year: 2017Title: Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma. Authors: Scott, J.S. / Degorce, S.L. / Anjum, R. / Culshaw, J. / Davies, R.D.M. / Davies, N.L. / Dillman, K.S. / Dowling, J.E. / Drew, L. / Ferguson, A.D. / Groombridge, S.D. / Halsall, C.T. / ...Authors: Scott, J.S. / Degorce, S.L. / Anjum, R. / Culshaw, J. / Davies, R.D.M. / Davies, N.L. / Dillman, K.S. / Dowling, J.E. / Drew, L. / Ferguson, A.D. / Groombridge, S.D. / Halsall, C.T. / Hudson, J.A. / Lamont, S. / Lindsay, N.A. / Marden, S.K. / Mayo, M.F. / Pease, J.E. / Perkins, D.R. / Pink, J.H. / Robb, G.R. / Rosen, A. / Shen, M. / McWhirter, C. / Wu, D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5k7g.cif.gz 5k7g.cif.gz | 460.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5k7g.ent.gz pdb5k7g.ent.gz | 380.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5k7g.json.gz 5k7g.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k7/5k7g https://data.pdbj.org/pub/pdb/validation_reports/k7/5k7g ftp://data.pdbj.org/pub/pdb/validation_reports/k7/5k7g ftp://data.pdbj.org/pub/pdb/validation_reports/k7/5k7g | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5k72C  5k75C  5k76C  5k7iC  2nruS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 33876.215 Da / Num. of mol.: 4 / Fragment: UNP residues 160-460 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: IRAK4 / Production host: Homo sapiens (human) / Gene: IRAK4 / Production host:  References: UniProt: Q9NWZ3, non-specific serine/threonine protein kinase #2: Chemical | ChemComp-SO4 / #3: Chemical | ChemComp-6R0 / ( #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.51 Å3/Da / Density % sol: 51.07 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 7.2 / Details: 2.5 M ammonium sulfate, 0.1 M Hepes / PH range: 6.8-7.2 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 17-ID / Wavelength: 1.10406 Å / Beamline: 17-ID / Wavelength: 1.10406 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Apr 27, 2015 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.10406 Å / Relative weight: 1 |
| Reflection | Resolution: 2.23→45.29 Å / Num. obs: 61883 / % possible obs: 95.8 % / Redundancy: 4 % / CC1/2: 0.995 / Rmerge(I) obs: 0.104 / Net I/σ(I): 10 |
| Reflection shell | Resolution: 2.23→2.24 Å / Redundancy: 3.4 % / Rmerge(I) obs: 0.685 / Mean I/σ(I) obs: 2.2 / % possible all: 89.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2NRU Resolution: 2.23→45.29 Å / Cor.coef. Fo:Fc: 0.925 / Cor.coef. Fo:Fc free: 0.908 / Rfactor Rfree error: 0 / SU R Cruickshank DPI: 0.288 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.279 / SU Rfree Blow DPI: 0.195 / SU Rfree Cruickshank DPI: 0.199
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 148.35 Å2 / Biso mean: 47.04 Å2 / Biso min: 18.4 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.23→45.29 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.23→2.29 Å / Total num. of bins used: 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller















 PDBj
PDBj