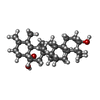
Entry Database : PDB / ID : 5k3mTitle Crystal structure of Retinoic acid receptor-related orphan receptor (ROR) gamma ligand binding domain complex with UUA Nuclear receptor ROR-gamma Keywords / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 2.894 Å Model details ROR gamma ligand binding domain complex with 444 Authors Huang, P. / Rastinejad, F. Journal : To be published Title : Crystal structure of Retinoic acid receptor-related orphan receptor (ROR) gamma ligand binding domain complex with UUAAuthors : Lu, J. / Rastinejad, F. History Deposition May 19, 2016 Deposition site / Processing site Revision 1.0 May 31, 2017 Provider / Type Revision 1.1 Mar 6, 2024 Group / Database references / Category / chem_comp_bond / database_2Item / _database_2.pdbx_database_accession
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.894 Å
MOLECULAR REPLACEMENT / Resolution: 2.894 Å  Authors
Authors Citation
Citation Journal: To be published
Journal: To be published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5k3m.cif.gz
5k3m.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5k3m.ent.gz
pdb5k3m.ent.gz PDB format
PDB format 5k3m.json.gz
5k3m.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/k3/5k3m
https://data.pdbj.org/pub/pdb/validation_reports/k3/5k3m ftp://data.pdbj.org/pub/pdb/validation_reports/k3/5k3m
ftp://data.pdbj.org/pub/pdb/validation_reports/k3/5k3m Links
Links Assembly
Assembly


 Components
Components Homo sapiens (human) / Gene: RORC, NR1F3, RORG, RZRG / Plasmid: pET28 / Production host:
Homo sapiens (human) / Gene: RORC, NR1F3, RORG, RZRG / Plasmid: pET28 / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  APS
APS  / Beamline: 19-ID / Wavelength: 0.9793 Å
/ Beamline: 19-ID / Wavelength: 0.9793 Å Processing
Processing MOLECULAR REPLACEMENT / Resolution: 2.894→34.67 Å / FOM work R set: 0.645 / Cross valid method: FREE R-VALUE / σ(F): 0 / Phase error: 40.91 / Stereochemistry target values: TWIN_LSQ_F
MOLECULAR REPLACEMENT / Resolution: 2.894→34.67 Å / FOM work R set: 0.645 / Cross valid method: FREE R-VALUE / σ(F): 0 / Phase error: 40.91 / Stereochemistry target values: TWIN_LSQ_F Movie
Movie Controller
Controller














 PDBj
PDBj