+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5jeu | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
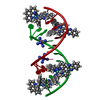
| Title | del-[Ru(phen)2(dppz)]2+ bound to d(TCGGCGCCGA) with Ba2+ | ||||||||||||||||||||
 Components Components | DNA (5'-D(* Keywords KeywordsDNA / ruthenium / light-switch / semi-intercalation | Function / homology | Delta-Ru(phen)2(dppz) complex / : / DNA |  Function and homology information Function and homology informationBiological species | synthetic construct (others) | Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 0.97 Å SAD / Resolution: 0.97 Å  Authors AuthorsHall, J.P. / Cardin, C.J. |  Citation Citation Journal: Nucleic Acids Res. / Year: 2016 Journal: Nucleic Acids Res. / Year: 2016Title: Delta chirality ruthenium 'light-switch' complexes can bind in the minor groove of DNA with five different binding modes. Authors: Hall, J.P. / Keane, P.M. / Beer, H. / Buchner, K. / Winter, G. / Sorensen, T.L. / Cardin, D.J. / Brazier, J.A. / Cardin, C.J. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5jeu.cif.gz 5jeu.cif.gz | 37 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5jeu.ent.gz pdb5jeu.ent.gz | 26.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5jeu.json.gz 5jeu.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  5jeu_validation.pdf.gz 5jeu_validation.pdf.gz | 1 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  5jeu_full_validation.pdf.gz 5jeu_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  5jeu_validation.xml.gz 5jeu_validation.xml.gz | 3.6 KB | Display | |
| Data in CIF |  5jeu_validation.cif.gz 5jeu_validation.cif.gz | 4.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/je/5jeu https://data.pdbj.org/pub/pdb/validation_reports/je/5jeu ftp://data.pdbj.org/pub/pdb/validation_reports/je/5jeu ftp://data.pdbj.org/pub/pdb/validation_reports/je/5jeu | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: DNA chain | Mass: 3045.992 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) | ||||
|---|---|---|---|---|---|
| #2: Chemical | ChemComp-BA / | ||||
| #3: Chemical | | #4: Chemical | ChemComp-CL / | #5: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.85 Å3/Da / Density % sol: 56.92 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / pH: 7 Details: 1uL 4mM del-[Ru(phen)2(dppz)]2+, 1uL 2mM d(TCGGCGCCGA), 6uL of a solution containing 40mM Na-cacodylate pH 7, 12mM spermine, 80mM KCl, 20mM BaCl2, 10% (v/v) hexylene glycol |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I02 / Wavelength: 0.8266 Å / Beamline: I02 / Wavelength: 0.8266 Å |
| Detector | Type: DECTRIS PILATUS 6M-F / Detector: PIXEL / Date: Dec 14, 2012 |
| Radiation | Monochromator: dual Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.8266 Å / Relative weight: 1 |
| Reflection | Resolution: 0.97→25.18 Å / Num. obs: 20581 / % possible obs: 96.7 % / Observed criterion σ(F): -3 / Redundancy: 6.4 % / CC1/2: 0.999 / Rmerge(I) obs: 0.027 / Net I/σ(I): 25.3 |
| Reflection shell | Resolution: 0.97→1 Å / Redundancy: 6.2 % / Rmerge(I) obs: 0.857 / Mean I/σ(I) obs: 1.9 / % possible all: 97.3 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  SAD / Resolution: 0.97→25.177 Å / SU ML: 0.08 / Cross valid method: THROUGHOUT / σ(F): 1.37 / Phase error: 13.45 / Stereochemistry target values: ML SAD / Resolution: 0.97→25.177 Å / SU ML: 0.08 / Cross valid method: THROUGHOUT / σ(F): 1.37 / Phase error: 13.45 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 0.97→25.177 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj








