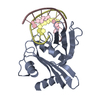
Entry Database : PDB / ID : 5idkTitle Crystal structure of West Nile Virus NS2B-NS3 protease in complex with a capped dipeptide boronate inhibitor Genome polyprotein,SERINE PROTEASE SUBUNIT NS2B, SERINE PROTEASE NS3 Keywords / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Method / / / Resolution : 1.5 Å Authors Hilgenfeld, R. / Zhang, L. Funding support Organization Grant number Country German Research Foundation KL-1356/3-1
Journal : J. Med. Chem. / Year : 2017Title : Peptide-Boronic Acid Inhibitors of Flaviviral Proteases: Medicinal Chemistry and Structural Biology.Authors : Nitsche, C. / Zhang, L. / Weigel, L.F. / Schilz, J. / Graf, D. / Bartenschlager, R. / Hilgenfeld, R. / Klein, C.D. History Deposition Feb 24, 2016 Deposition site / Processing site Revision 1.0 Dec 14, 2016 Provider / Type Revision 1.1 Dec 28, 2016 Group Revision 1.2 Jan 25, 2017 Group Revision 1.3 Jul 5, 2017 Group / Category / Item Revision 1.4 Sep 6, 2017 Group / Category / Item Revision 1.5 Jan 10, 2024 Group Advisory / Data collection ... Advisory / Data collection / Database references / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_unobs_or_zero_occ_atoms / struct_ncs_dom_lim Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id Revision 1.6 Oct 23, 2024 Group / Category / pdbx_modification_feature
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information West Nile virus
West Nile virus X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.5 Å
MOLECULAR REPLACEMENT / Resolution: 1.5 Å  Authors
Authors Germany, 1items
Germany, 1items  Citation
Citation Journal: J. Med. Chem. / Year: 2017
Journal: J. Med. Chem. / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5idk.cif.gz
5idk.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5idk.ent.gz
pdb5idk.ent.gz PDB format
PDB format 5idk.json.gz
5idk.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/id/5idk
https://data.pdbj.org/pub/pdb/validation_reports/id/5idk ftp://data.pdbj.org/pub/pdb/validation_reports/id/5idk
ftp://data.pdbj.org/pub/pdb/validation_reports/id/5idk
 Links
Links Assembly
Assembly



 Components
Components West Nile virus / Production host:
West Nile virus / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  PETRA III, DESY
PETRA III, DESY  / Beamline: P11 / Wavelength: 0.9919 Å
/ Beamline: P11 / Wavelength: 0.9919 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller











 PDBj
PDBj








