Entry Database : PDB / ID : 5d5lTitle PreQ1-II riboswitch with an engineered G-U wobble pair bound to Cs+ PreQ1-II riboswitch Keywords / / / Function / homology / / / Biological species Lactobacillus rhamnosus LOCK908 (bacteria)Method / / / / Resolution : 2.5 Å Authors Wedekind, J.E. / Liberman, J.A. / Salim, M. Funding support Organization Grant number Country National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) GM063162 National Institutes of Health/National Center for Research Resources (NIH/NCRR) RR026501
Journal : to be published Title : Cesium(I) binding to G-U-wobble base pairs in preQ1 riboswitches with implications for crystallographic phasingAuthors : Belashov, I.A. / Liberman, J.A. / Salim, M. / Wedekind, J.E. History Deposition Aug 10, 2015 Deposition site / Processing site Revision 1.0 Aug 10, 2016 Provider / Type Revision 1.1 Sep 20, 2017 Group / Derived calculations / Category / pdbx_struct_oper_listItem / _pdbx_struct_oper_list.symmetry_operationRevision 1.2 Dec 4, 2019 Group / Category / Item Revision 1.3 Sep 27, 2023 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn / struct_conn_type Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr1_symmetry / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_auth_comp_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr2_label_atom_id / _pdbx_struct_conn_angle.ptnr2_label_comp_id / _pdbx_struct_conn_angle.ptnr2_symmetry / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.ptnr3_symmetry / _pdbx_struct_conn_angle.value / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr1_symmetry / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn.ptnr2_symmetry / _struct_conn_type.id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Lactobacillus rhamnosus LOCK908 (bacteria)
Lactobacillus rhamnosus LOCK908 (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.5 Å
molecular replacement / Resolution: 2.5 Å  Authors
Authors United States, 2items
United States, 2items  Citation
Citation Journal: to be published
Journal: to be published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5d5l.cif.gz
5d5l.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5d5l.ent.gz
pdb5d5l.ent.gz PDB format
PDB format 5d5l.json.gz
5d5l.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/d5/5d5l
https://data.pdbj.org/pub/pdb/validation_reports/d5/5d5l ftp://data.pdbj.org/pub/pdb/validation_reports/d5/5d5l
ftp://data.pdbj.org/pub/pdb/validation_reports/d5/5d5l
 Links
Links Assembly
Assembly

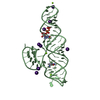
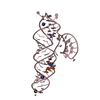
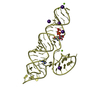
 Components
Components Lactobacillus rhamnosus LOCK908 (bacteria)
Lactobacillus rhamnosus LOCK908 (bacteria) X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SSRL
SSRL  / Beamline: BL11-1 / Wavelength: 1.17 Å
/ Beamline: BL11-1 / Wavelength: 1.17 Å molecular replacement
molecular replacement Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller











 PDBj
PDBj