+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5cl1 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Complex structure of Norrin with human Frizzled 4 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SIGNALING PROTEIN / Wnt / Norrin / Frizzled | ||||||
| Function / homology |  Function and homology information Function and homology informationcerebellum vasculature morphogenesis / Wnt signaling pathway, calcium modulating pathway / retina blood vessel maintenance / cone retinal bipolar cell differentiation / Norrin signaling pathway / extracellular matrix-cell signaling / progesterone secretion / retinal rod cell differentiation / re-entry into mitotic cell cycle / retinal blood vessel morphogenesis ...cerebellum vasculature morphogenesis / Wnt signaling pathway, calcium modulating pathway / retina blood vessel maintenance / cone retinal bipolar cell differentiation / Norrin signaling pathway / extracellular matrix-cell signaling / progesterone secretion / retinal rod cell differentiation / re-entry into mitotic cell cycle / retinal blood vessel morphogenesis / retina vasculature morphogenesis in camera-type eye / locomotion involved in locomotory behavior / Signaling by RNF43 mutants / WNT5A-dependent internalization of FZD4 / glycine metabolic process / regulation of vascular endothelial growth factor receptor signaling pathway / retina layer formation / positive regulation of neuron projection arborization / Wnt receptor activity / retinal pigment epithelium development / non-canonical Wnt signaling pathway / microglial cell proliferation / endothelial cell differentiation / Wnt-protein binding / dendritic spine development / establishment of blood-retinal barrier / vacuole organization / microglia differentiation / protein targeting to lysosome / establishment of blood-brain barrier / frizzled binding / optic nerve development / positive regulation of dendrite morphogenesis / Class B/2 (Secretin family receptors) / cytokine receptor activity / retinal ganglion cell axon guidance / lens development in camera-type eye / ubiquitin-dependent endocytosis / smoothened signaling pathway / cytokine binding / exploration behavior / decidualization / action potential / negative regulation of cell-substrate adhesion / carbohydrate transmembrane transporter activity / blood vessel remodeling / maltose binding / maltose transport / canonical Wnt signaling pathway / maltodextrin transmembrane transport / response to axon injury / vasculogenesis / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / tricarboxylic acid cycle / cellular response to retinoic acid / glutathione metabolic process / visual perception / substrate adhesion-dependent cell spreading / transforming growth factor beta receptor signaling pathway / Regulation of FZD by ubiquitination / cytokine activity / cellular response to leukemia inhibitory factor / PDZ domain binding / Asymmetric localization of PCP proteins / clathrin-coated endocytic vesicle membrane / sensory perception of sound / : / G protein-coupled receptor activity / Wnt signaling pathway / neuron differentiation / cell-cell junction / nervous system development / mitotic cell cycle / Cargo recognition for clathrin-mediated endocytosis / signaling receptor activity / amyloid-beta binding / Clathrin-mediated endocytosis / outer membrane-bounded periplasmic space / neuron apoptotic process / Ca2+ pathway / angiogenesis / cellular response to hypoxia / transcription by RNA polymerase II / response to hypoxia / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / cell population proliferation / cilium / positive regulation of cell migration / protein ubiquitination / inflammatory response / protein heterodimerization activity / ubiquitin protein ligase binding / dendrite / positive regulation of DNA-templated transcription / protein-containing complex binding / glutamatergic synapse / cell surface / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / extracellular space Similarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 3.8 Å molecular replacement / Resolution: 3.8 Å | ||||||
 Authors Authors | Wang, Z. / Ke, J. / Shen, G. / Cheng, Z. / Xu, H.E. / Xu, W. | ||||||
 Citation Citation |  Journal: Cell Res. / Year: 2015 Journal: Cell Res. / Year: 2015Title: Structural basis of the Norrin-Frizzled 4 interaction. Authors: Shen, G. / Ke, J. / Wang, Z. / Cheng, Z. / Gu, X. / Wei, Y. / Melcher, K. / Xu, H.E. / Xu, W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5cl1.cif.gz 5cl1.cif.gz | 462 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5cl1.ent.gz pdb5cl1.ent.gz | 385.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5cl1.json.gz 5cl1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cl/5cl1 https://data.pdbj.org/pub/pdb/validation_reports/cl/5cl1 ftp://data.pdbj.org/pub/pdb/validation_reports/cl/5cl1 ftp://data.pdbj.org/pub/pdb/validation_reports/cl/5cl1 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 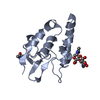 5cm4C  4my2S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: _ / Refine code: _
NCS ensembles :
|
- Components
Components
| #1: Protein | Mass: 53664.043 Da / Num. of mol.: 2 / Fragment: UNP Q00604 residues 31-133 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) Homo sapiens (human)Gene: malE, Z5632, ECs5017, NDP, EVR2 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: P0AEY0, UniProt: Q00604 Trichoplusia ni (cabbage looper) / References: UniProt: P0AEY0, UniProt: Q00604#2: Protein | Mass: 15098.341 Da / Num. of mol.: 2 / Fragment: UNP residues 38-160 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: FZD4 / Production host: Homo sapiens (human) / Gene: FZD4 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: Q9ULV1 Trichoplusia ni (cabbage looper) / References: UniProt: Q9ULV1#3: Sugar | ChemComp-NAG / Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.62 Å3/Da / Density % sol: 66.03 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: 200 mM ammonium sulfate, 100 mM sodium cacodylate trihydrate (pH 6.5), 15% w/v polyethylene glycol 8,000 |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 5.0.1 / Wavelength: 1 Å / Beamline: 5.0.1 / Wavelength: 1 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: May 29, 2015 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: Single crystal, cylindrically bent, Si(220) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 3.8→50 Å / Num. obs: 18303 / % possible obs: 90.5 % / Redundancy: 5.9 % / Rmerge(I) obs: 0.264 / Rpim(I) all: 0.117 / Rrim(I) all: 0.28 / Χ2: 1.123 / Net I/av σ(I): 4.476 / Net I/σ(I): 3.5 / Num. measured all: 107966 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1 / Rejects: _
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4MY2 Resolution: 3.8→50 Å / Cor.coef. Fo:Fc: 0.879 / Cor.coef. Fo:Fc free: 0.808 / SU B: 144.371 / SU ML: 0.843 / Cross valid method: THROUGHOUT / ESU R Free: 0.905 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : WITH TLS ADDED
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 195.82 Å2 / Biso mean: 112.326 Å2 / Biso min: 71.47 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 3.8→50 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | Refine-ID: X-RAY DIFFRACTION / Type: interatomic distance / Weight position: 0.05
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.8→3.892 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj











