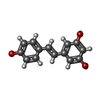
登録情報 データベース : PDB / ID : 5btrタイトル Crystal structure of SIRT1 in complex with resveratrol and an AMC-containing peptide AMC-containing peptide NAD-dependent protein deacetylase sirtuin-1 キーワード / / / / / / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト)synthetic (人工物) 手法 / / / 解像度 : 3.2 Å データ登録者 Cao, D. / Wang, M. / Qiu, X. / Liu, D. / Jiang, H. / Yang, N. / Xu, R.M. 資金援助 組織 認可番号 国 Ministry of Science and Technology (China) 2012CB910702 National Natural Science Foundation of China 31430018 National Natural Science Foundation of China 31210103914 Strategic Priority Research Program of Chinese Academy of Sciences XDB08010100 Key Research Program of Chinese Academy of Sciences KJZD-EW-L05 National Key New Drug Creation and Manufacturing Program of China 2014ZX09507002
ジャーナル : Genes Dev. / 年 : 2015タイトル : Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol著者 : Cao, D. / Wang, M. / Qiu, X. / Liu, D. / Jiang, H. / Yang, N. / Xu, R.M. 履歴 登録 2015年6月3日 登録サイト / 処理サイト 改定 1.0 2015年7月8日 Provider / タイプ 改定 1.1 2016年12月28日 Group 改定 1.2 2017年10月18日 Group / カテゴリ / Item 改定 1.3 2023年11月8日 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description カテゴリ chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_conn / struct_conn_type Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn_type.id
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 3.2 Å
分子置換 / 解像度: 3.2 Å  データ登録者
データ登録者 中国, 6件
中国, 6件  引用
引用 ジャーナル: Genes Dev. / 年: 2015
ジャーナル: Genes Dev. / 年: 2015 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 5btr.cif.gz
5btr.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb5btr.ent.gz
pdb5btr.ent.gz PDB形式
PDB形式 5btr.json.gz
5btr.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 5btr_validation.pdf.gz
5btr_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 5btr_full_validation.pdf.gz
5btr_full_validation.pdf.gz 5btr_validation.xml.gz
5btr_validation.xml.gz 5btr_validation.cif.gz
5btr_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/bt/5btr
https://data.pdbj.org/pub/pdb/validation_reports/bt/5btr ftp://data.pdbj.org/pub/pdb/validation_reports/bt/5btr
ftp://data.pdbj.org/pub/pdb/validation_reports/bt/5btr
 リンク
リンク 集合体
集合体



 要素
要素 Homo sapiens (ヒト) / 遺伝子: SIRT1, SIR2L1 / プラスミド: pET-28a smt3 / 発現宿主:
Homo sapiens (ヒト) / 遺伝子: SIRT1, SIR2L1 / プラスミド: pET-28a smt3 / 発現宿主: 
 X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  SSRF
SSRF  / ビームライン: BL17U / 波長: 0.9792 Å
/ ビームライン: BL17U / 波長: 0.9792 Å 分子置換
分子置換 解析
解析 分子置換
分子置換 ムービー
ムービー コントローラー
コントローラー













 PDBj
PDBj