+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5a9i | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the extracellular domain of PepT2 | ||||||
 Components Components | SOLUTE CARRIER FAMILY 15 MEMBER 2 | ||||||
 Keywords Keywords | TRANSPORT PROTEIN / PEPT2 / EXTRACELLULAR DOMAIN / MFS | ||||||
| Function / homology |  Function and homology information Function and homology informationhigh-affinity oligopeptide transmembrane transporter activity / tripeptide import across plasma membrane / oligopeptide transport / dipeptide transport / peptidoglycan transport / dipeptide import across plasma membrane / tripeptide transmembrane transporter activity / metanephric proximal tubule development / peptide:proton symporter activity / antibacterial innate immune response ...high-affinity oligopeptide transmembrane transporter activity / tripeptide import across plasma membrane / oligopeptide transport / dipeptide transport / peptidoglycan transport / dipeptide import across plasma membrane / tripeptide transmembrane transporter activity / metanephric proximal tubule development / peptide:proton symporter activity / antibacterial innate immune response / dipeptide transmembrane transporter activity / regulation of nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway / xenobiotic transport / renal absorption / phagocytic vesicle membrane / protein transport / apical plasma membrane / membrane / plasma membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 2.84 Å SAD / Resolution: 2.84 Å | ||||||
 Authors Authors | Beale, J.H. / Newstead, S. | ||||||
 Citation Citation |  Journal: Structure / Year: 2015 Journal: Structure / Year: 2015Title: Crystal Structures of the Extracellular Domain from Pept1 and Pept2 Provide Novel Insights Into Mammalian Peptide Transport Authors: Beale, J.H. / Parker, J.L. / Samsudin, F. / Barrett, A.L. / Senan, A. / Bird, L.E. / Scott, D. / Owens, R.J. / Sanson, M.S.P. / Tucker, S.J. / Meredith, D. / Fowler, P.W. / Newstead, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5a9i.cif.gz 5a9i.cif.gz | 239.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5a9i.ent.gz pdb5a9i.ent.gz | 197.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5a9i.json.gz 5a9i.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/a9/5a9i https://data.pdbj.org/pub/pdb/validation_reports/a9/5a9i ftp://data.pdbj.org/pub/pdb/validation_reports/a9/5a9i ftp://data.pdbj.org/pub/pdb/validation_reports/a9/5a9i | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 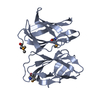
| ||||||||||||
| 2 | 
| ||||||||||||
| 3 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
| #1: Protein | Mass: 21694.936 Da / Num. of mol.: 3 / Fragment: EXTRACELLULAR DOMAIN, RESIDUES 410-601 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Chemical | #3: Chemical | ChemComp-GOL / | #4: Water | ChemComp-HOH / | Has protein modification | Y | Nonpolymer details | CITRATE (CIT): FROM CRYSTALLIS | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.47 Å3/Da / Density % sol: 64 % / Description: NONE |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop Details: 0.2 M (NH4)3 CITRATE PH 5.8, 21 % PEG 3350. 10 MG.ML AT 20 DEGREES CELCIUS |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04 / Wavelength: 0.97949 / Beamline: I04 / Wavelength: 0.97949 |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: May 6, 2013 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97949 Å / Relative weight: 1 |
| Reflection | Resolution: 2.81→58 Å / Num. obs: 21317 / % possible obs: 99.4 % / Observed criterion σ(I): 2 / Redundancy: 14 % / Biso Wilson estimate: 86.74 Å2 / CC1/2: 0.999 / Rmerge(I) obs: 0.15 / Net I/σ(I): 12.7 |
| Reflection shell | Resolution: 2.81→2.96 Å / Redundancy: 10 % / Rmerge(I) obs: 1.04 / Mean I/σ(I) obs: 2.3 / CC1/2: 0.697 / % possible all: 69.7 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  SAD SADStarting model: NONE Resolution: 2.84→47.87 Å / Cor.coef. Fo:Fc: 0.9371 / Cor.coef. Fo:Fc free: 0.898 / SU R Cruickshank DPI: 0.733 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.76 / SU Rfree Blow DPI: 0.324 / SU Rfree Cruickshank DPI: 0.328 Details: IDEAL-DIST CONTACT TERM CONTACT SETUP. ALL ATOMS HAVE CCP4 ATOM TYPE FROM LIBRARY.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 75.13 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.465 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.84→47.87 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.84→2.98 Å / Total num. of bins used: 11
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller















 PDBj
PDBj







