[English] 日本語
 Yorodumi
Yorodumi- PDB-4q99: Crystal structure of 2-mercapto-4-methylphenol bound to human car... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4q99 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of 2-mercapto-4-methylphenol bound to human carbonic anhydrase II | ||||||
 Components Components | Carbonic anhydrase 2 | ||||||
 Keywords Keywords | LYASE | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of dipeptide transmembrane transport / positive regulation of cellular pH reduction / Developmental Lineage of Pancreatic Ductal Cells / regulation of monoatomic anion transport / secretion / cyanamide hydratase / cyanamide hydratase activity / arylesterase activity / regulation of chloride transport / Reversible hydration of carbon dioxide ...positive regulation of dipeptide transmembrane transport / positive regulation of cellular pH reduction / Developmental Lineage of Pancreatic Ductal Cells / regulation of monoatomic anion transport / secretion / cyanamide hydratase / cyanamide hydratase activity / arylesterase activity / regulation of chloride transport / Reversible hydration of carbon dioxide / morphogenesis of an epithelium / angiotensin-activated signaling pathway / positive regulation of synaptic transmission, GABAergic / regulation of intracellular pH / carbonic anhydrase / carbonate dehydratase activity / carbon dioxide transport / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen / neuron cellular homeostasis / apical part of cell / myelin sheath / extracellular exosome / zinc ion binding / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.5 Å MOLECULAR REPLACEMENT / Resolution: 1.5 Å | ||||||
 Authors Authors | Martin, D.P. / Cohen, S.M. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2014 Journal: J.Med.Chem. / Year: 2014Title: Exploring the influence of the protein environment on metal-binding pharmacophores. Authors: Martin, D.P. / Blachly, P.G. / McCammon, J.A. / Cohen, S.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4q99.cif.gz 4q99.cif.gz | 71.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4q99.ent.gz pdb4q99.ent.gz | 51.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4q99.json.gz 4q99.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/q9/4q99 https://data.pdbj.org/pub/pdb/validation_reports/q9/4q99 ftp://data.pdbj.org/pub/pdb/validation_reports/q9/4q99 ftp://data.pdbj.org/pub/pdb/validation_reports/q9/4q99 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4q7pC  4q7sC  4q7vC  4q7wC  4q81C  4q83C  4q87C  4q8xC  4q8yC  4q8zC  4q90C  4q9yC  3ks3S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 29289.062 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CA2 / Production host: Homo sapiens (human) / Gene: CA2 / Production host:  |
|---|
-Non-polymers , 6 types, 192 molecules 

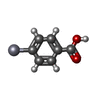








| #2: Chemical | ChemComp-ZN / |
|---|---|
| #3: Chemical | ChemComp-HT4 / |
| #4: Chemical | ChemComp-MBO / |
| #5: Chemical | ChemComp-GOL / |
| #6: Chemical | ChemComp-DMS / |
| #7: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.07 Å3/Da / Density % sol: 40.45 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion / pH: 8 Details: Ammonium sulfate, pH 8.0, VAPOR DIFFUSION, temperature 291K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: ENRAF-NONIUS FR591 / Wavelength: 1.5478 Å ROTATING ANODE / Type: ENRAF-NONIUS FR591 / Wavelength: 1.5478 Å |
| Detector | Type: BRUKER SMART 6000 / Detector: CCD / Date: Apr 28, 2014 |
| Radiation | Monochromator: Montels mirrors / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5478 Å / Relative weight: 1 |
| Reflection | Resolution: 1.5→30.43 Å / Num. all: 36641 / Num. obs: 36454 / % possible obs: 99.49 % / Observed criterion σ(F): 1 / Observed criterion σ(I): 1 |
| Reflection shell | Resolution: 1.5→1.539 Å / % possible all: 98.31 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 3KS3 Resolution: 1.5→30.43 Å / Cor.coef. Fo:Fc: 0.959 / Cor.coef. Fo:Fc free: 0.951 / SU B: 1.197 / SU ML: 0.045 / Cross valid method: THROUGHOUT / σ(F): 1 / ESU R: 0.073 / ESU R Free: 0.074 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 10.959 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.5→30.43 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller























 PDBj
PDBj





