| Entry | Database: PDB / ID: 4lbf
|
|---|
| Title | Crystal structure of HUMAN ALPHA-DEFENSIN 1 (HNP1) I20A/L25A mutant |
|---|
 Components Components | Neutrophil defensin 1 |
|---|
 Keywords Keywords | ANTIMICROBIAL PROTEIN / ANTIMICROBIAL PEPTIDE / HUMAN ALPHA DEFENSIN 1 / HUMAN NEUTROPHIL PEPTIDE 1 / ANTIBIOTIC / ANTIVIRAL DEFENSE / DEFENSIN / DISULFIDE BOND / FUNGICIDE / PHOSPHOPROTEIN / SECRETED |
|---|
| Function / homology |  Function and homology information Function and homology information
disruption of plasma membrane integrity in another organism / pore-forming activity / Defensins / : / T cell chemotaxis / Alpha-defensins / defense response to protozoan / defense response to fungus / estrogen receptor signaling pathway / innate immune response in mucosa ...disruption of plasma membrane integrity in another organism / pore-forming activity / Defensins / : / T cell chemotaxis / Alpha-defensins / defense response to protozoan / defense response to fungus / estrogen receptor signaling pathway / innate immune response in mucosa / Golgi lumen / : / chemotaxis / azurophil granule lumen / antimicrobial humoral immune response mediated by antimicrobial peptide / antibacterial humoral response / cellular response to lipopolysaccharide / defense response to virus / killing of cells of another organism / defense response to Gram-negative bacterium / immune response / defense response to Gram-positive bacterium / Neutrophil degranulation / extracellular space / extracellular exosome / extracellular regionSimilarity search - Function Mammalian defensins signature. / Alpha-defensin, C-terminal / Mammalian defensin / Alpha-defensin propeptide / Alpha-defensin / Defensin propeptide / Defensin propeptide / Beta/alpha-defensin, C-terminal / Defensin/corticostatin familySimilarity search - Domain/homology |
|---|
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.7 Å MOLECULAR REPLACEMENT / Resolution: 1.7 Å |
|---|
 Authors Authors | Tolbert, W.D. / Wu, X. / Pazgier, M. |
|---|
 Citation Citation |  Journal: Plos One / Year: 2013 Journal: Plos One / Year: 2013
Title: Single, Double and Quadruple Alanine Substitutions at Oligomeric Interfaces Identify Hydrophobicity as the Key Determinant of Human Neutrophil Alpha Defensin HNP1 Function.
Authors: Zhao, L. / Tolbert, W.D. / Ericksen, B. / Zhan, C. / Wu, X. / Yuan, W. / Li, X. / Pazgier, M. / Lu, W. |
|---|
| History | | Deposition | Jun 20, 2013 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Nov 27, 2013 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Sep 20, 2023 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_ref_seq_dif / struct_site
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ref_seq_dif.details / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id |
|---|
| Revision 1.2 | Nov 6, 2024 | Group: Structure summary / Category: pdbx_entry_details / pdbx_modification_feature |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.7 Å
MOLECULAR REPLACEMENT / Resolution: 1.7 Å  Authors
Authors Citation
Citation Journal: Plos One / Year: 2013
Journal: Plos One / Year: 2013 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4lbf.cif.gz
4lbf.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4lbf.ent.gz
pdb4lbf.ent.gz PDB format
PDB format 4lbf.json.gz
4lbf.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/lb/4lbf
https://data.pdbj.org/pub/pdb/validation_reports/lb/4lbf ftp://data.pdbj.org/pub/pdb/validation_reports/lb/4lbf
ftp://data.pdbj.org/pub/pdb/validation_reports/lb/4lbf
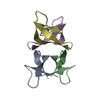


 Links
Links Assembly
Assembly


 Components
Components Homo sapiens (human) / References: UniProt: P59665
Homo sapiens (human) / References: UniProt: P59665 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.54 Å
ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.54 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller




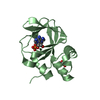



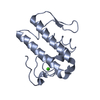

 PDBj
PDBj




