[English] 日本語
 Yorodumi
Yorodumi- PDB-4d09: PDE2a catalytic domain in complex with a brain penetrant inhibitor -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4d09 | ||||||
|---|---|---|---|---|---|---|---|
| Title | PDE2a catalytic domain in complex with a brain penetrant inhibitor | ||||||
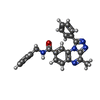
 Components Components | CGMP-DEPENDENT 3', 5'-CYCLIC PHOSPHODIESTERASE | ||||||
 Keywords Keywords | HYDROLASE / CYCLIC NUCLEOTIDE PHOSPHODIESTERASES / TYPE 2 / TYPE 4 / DISEASE MODELS / ANIMAL / DRUG EVALUATION / PRECLINICAL / PHOSPHODIESTERASE 2 INHIBITORS / PHOSPHODIESTERASE INHIBITORS / PROTEIN BINDING / STRUCTURE-ACTIVITY RELATIONSHIP | ||||||
| Function / homology |  Function and homology information Function and homology information: / cellular response to 2,3,7,8-tetrachlorodibenzodioxine / cellular response to macrophage colony-stimulating factor stimulus / cellular response to cGMP / negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway / positive regulation of vascular permeability / heart valve development / cellular response to granulocyte macrophage colony-stimulating factor stimulus / negative regulation of vascular permeability / regulation of mitochondrion organization ...: / cellular response to 2,3,7,8-tetrachlorodibenzodioxine / cellular response to macrophage colony-stimulating factor stimulus / cellular response to cGMP / negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway / positive regulation of vascular permeability / heart valve development / cellular response to granulocyte macrophage colony-stimulating factor stimulus / negative regulation of vascular permeability / regulation of mitochondrion organization / establishment of endothelial barrier / : / 3',5'-cGMP-stimulated cyclic-nucleotide phosphodiesterase activity / : / 3',5'-cyclic-nucleotide phosphodiesterase / negative regulation of receptor guanylyl cyclase signaling pathway / ventricular septum development / aorta development / cGMP catabolic process / cGMP effects / phosphate ion binding / TPR domain binding / cGMP binding / 3',5'-cyclic-GMP phosphodiesterase activity / 3',5'-cyclic-AMP phosphodiesterase activity / monocyte differentiation / cellular response to transforming growth factor beta stimulus / cAMP binding / cellular response to cAMP / synaptic membrane / cellular response to mechanical stimulus / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / cellular response to xenobiotic stimulus / positive regulation of inflammatory response / presynaptic membrane / G alpha (s) signalling events / mitochondrial outer membrane / mitochondrial inner membrane / mitochondrial matrix / positive regulation of gene expression / perinuclear region of cytoplasm / magnesium ion binding / negative regulation of transcription by RNA polymerase II / endoplasmic reticulum / Golgi apparatus / protein homodimerization activity / zinc ion binding / identical protein binding / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.5 Å MOLECULAR REPLACEMENT / Resolution: 2.5 Å | ||||||
 Authors Authors | Buijnsters, P. / Andres, J.I. / DeAngelis, M. / Langlois, X. / Rombouts, F. / Sanderson, W. / Tresadern, G. / Trabanco, A. / VanHoof, G. / VanRoosbroeck, Y. | ||||||
 Citation Citation |  Journal: Acs Med.Chem.Lett. / Year: 2014 Journal: Acs Med.Chem.Lett. / Year: 2014Title: Structure-Based Design of a Potent, Selective, and Brain Penetrating Pde2 Inhibitor with Demonstrated Target Engagement. Authors: Buijnsters, P. / De Angelis, M. / Langlois, X. / Rombouts, F.J.R. / Sanderson, W. / Tresadern, G. / Ritchie, A. / Trabanco, A.A. / Vanhoof, G. / Roosbroeck, Y.V. / Andres, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4d09.cif.gz 4d09.cif.gz | 278.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4d09.ent.gz pdb4d09.ent.gz | 226.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4d09.json.gz 4d09.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/d0/4d09 https://data.pdbj.org/pub/pdb/validation_reports/d0/4d09 ftp://data.pdbj.org/pub/pdb/validation_reports/d0/4d09 ftp://data.pdbj.org/pub/pdb/validation_reports/d0/4d09 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 41432.375 Da / Num. of mol.: 4 / Fragment: CATALYTIC DOMAIN, RESIDUES 578-921 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Production host: HOMO SAPIENS (human) / Production host:  References: UniProt: O00408, 3',5'-cyclic-nucleotide phosphodiesterase #2: Chemical | ChemComp-788 / #3: Chemical | ChemComp-ZN / #4: Chemical | ChemComp-MG / #5: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.3 Å3/Da / Density % sol: 46.4 % / Description: NONE |
|---|---|
| Crystal grow | pH: 7 Details: 0.2 M MGCL2, 0.1 M TRIS-HCL PH 7.0 AND 20 % PEG 3350 IN PRESENCE OF 1 MM LIGAND |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.54 ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.54 |
| Detector | Type: RIGAKU SATURN 944 / Detector: CCD / Date: Jun 8, 2009 / Details: VARIAMAXHF |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.54 Å / Relative weight: 1 |
| Reflection | Resolution: 2.5→30 Å / Num. obs: 44706 / % possible obs: 93.7 % / Observed criterion σ(I): 1.8 / Redundancy: 1.9 % / Rmerge(I) obs: 0.1 / Net I/σ(I): 5.8 |
| Reflection shell | Resolution: 2.5→2.6 Å / Redundancy: 1.9 % / Rmerge(I) obs: 0.38 / Mean I/σ(I) obs: 1.8 / % possible all: 85.4 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: INTERNAL PDE2A MODEL Resolution: 2.5→19.77 Å / Cor.coef. Fo:Fc: 0.924 / Cor.coef. Fo:Fc free: 0.858 / SU B: 13.952 / SU ML: 0.303 / Cross valid method: THROUGHOUT / ESU R Free: 0.369 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 30.798 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.5→19.77 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj