| Entry | Database: PDB / ID: 4c9y
|
|---|
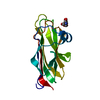
| Title | Structural Basis for the microtubule binding of the human kinetochore Ska complex |
|---|
 Components Components | SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 1 |
|---|
 Keywords Keywords | CELL CYCLE / CELL DIVISON / KINETOCHORE-MICROTUBULE ATTACHMENT / WINGED-HELIX DOMAIN |
|---|
| Function / homology |  Function and homology information Function and homology information
negative regulation of mitotic spindle assembly checkpoint signaling / SKA complex / establishment of meiotic spindle orientation / metaphase chromosome alignment / mitotic spindle microtubule / outer kinetochore / attachment of mitotic spindle microtubules to kinetochore / spindle assembly involved in female meiosis / mitotic metaphase chromosome alignment / mitotic sister chromatid segregation ...negative regulation of mitotic spindle assembly checkpoint signaling / SKA complex / establishment of meiotic spindle orientation / metaphase chromosome alignment / mitotic spindle microtubule / outer kinetochore / attachment of mitotic spindle microtubules to kinetochore / spindle assembly involved in female meiosis / mitotic metaphase chromosome alignment / mitotic sister chromatid segregation / regulation of microtubule polymerization or depolymerization / intercellular bridge / positive regulation of microtubule polymerization / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / Resolution of Sister Chromatid Cohesion / spindle microtubule / chromosome segregation / RHO GTPases Activate Formins / kinetochore / centriolar satellite / mitotic spindle / Separation of Sister Chromatids / mitotic cell cycle / microtubule cytoskeleton / microtubule binding / cilium / ciliary basal body / cell division / centrosome / nucleoplasm / cytosolSimilarity search - Function Ska1 microtubule binding domain-like / Spindle and kinetochore-associated protein 1 / SKA1 microtubule binding domain / Spindle and kinetochore-associated protein 1 / Arc Repressor Mutant, subunit A / Orthogonal Bundle / Mainly AlphaSimilarity search - Domain/homology |
|---|
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 2.01 Å SAD / Resolution: 2.01 Å |
|---|
 Authors Authors | Abad, M. / Medina, B. / Santamaria, A. / Zou, J. / Plasberg-Hill, C. / Madhumalar, A. / Jayachandran, U. / Redli, P.M. / Rappsilber, J. / Nigg, E.A. / Jeyaprakash, A.A. |
|---|
 Citation Citation |  Journal: Nat.Commun. / Year: 2014 Journal: Nat.Commun. / Year: 2014
Title: Structural Basis for Microtubule Recognition by the Human Kinetochore Ska Complex.
Authors: Abad, M.A. / Medina, B. / Santamaria, A. / Zou, J. / Plasberg-Hill, C. / Madhumalar, A. / Jayachandran, U. / Redli, P.M. / Rappsilber, J. / Nigg, E.A. / Jeyaprakash, A.A. |
|---|
| History | | Deposition | Oct 4, 2013 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Jan 22, 2014 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Nov 20, 2024 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Other / Structure summary
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_database_status / pdbx_entry_details / pdbx_modification_feature / struct_conn
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_sf / _pdbx_entry_details.has_protein_modification / _struct_conn.pdbx_leaving_atom_flag |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information HOMO SAPIENS (human)
HOMO SAPIENS (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  SAD / Resolution: 2.01 Å
SAD / Resolution: 2.01 Å  Authors
Authors Citation
Citation Journal: Nat.Commun. / Year: 2014
Journal: Nat.Commun. / Year: 2014 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4c9y.cif.gz
4c9y.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4c9y.ent.gz
pdb4c9y.ent.gz PDB format
PDB format 4c9y.json.gz
4c9y.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/c9/4c9y
https://data.pdbj.org/pub/pdb/validation_reports/c9/4c9y ftp://data.pdbj.org/pub/pdb/validation_reports/c9/4c9y
ftp://data.pdbj.org/pub/pdb/validation_reports/c9/4c9y Links
Links Assembly
Assembly


 Components
Components HOMO SAPIENS (human) / Plasmid: PEC-CDF-HIS / Production host:
HOMO SAPIENS (human) / Plasmid: PEC-CDF-HIS / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I02 / Wavelength: 1.28
/ Beamline: I02 / Wavelength: 1.28  Processing
Processing SAD
SAD Movie
Movie Controller
Controller











 PDBj
PDBj



