[English] 日本語
 Yorodumi
Yorodumi- PDB-4bz7: Crystal structure of Schistosoma mansoni HDAC8 complexed with M344 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4bz7 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of Schistosoma mansoni HDAC8 complexed with M344 | ||||||
 Components Components | HISTONE DEACETYLASE 8 | ||||||
 Keywords Keywords | HYDROLASE / PLATYHELMINTHS / INHIBITION | ||||||
| Function / homology |  Function and homology information Function and homology informationhistone deacetylase / histone deacetylase activity / heterochromatin formation / metal ion binding / nucleus Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.65 Å MOLECULAR REPLACEMENT / Resolution: 1.65 Å | ||||||
 Authors Authors | Marek, M. / Romier, C. | ||||||
 Citation Citation |  Journal: Plos Pathog. / Year: 2013 Journal: Plos Pathog. / Year: 2013Title: Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni. Authors: Marek, M. / Kannan, S. / Hauser, A. / Moraes Mourao, M. / Caby, S. / Cura, V. / Stolfa, D.A. / Schmidtkunz, K. / Lancelot, J. / Andrade, L. / Renaud, J. / Oliveira, G. / Sippl, W. / Jung, M. ...Authors: Marek, M. / Kannan, S. / Hauser, A. / Moraes Mourao, M. / Caby, S. / Cura, V. / Stolfa, D.A. / Schmidtkunz, K. / Lancelot, J. / Andrade, L. / Renaud, J. / Oliveira, G. / Sippl, W. / Jung, M. / Cavarelli, J. / Pierce, R.J. / Romier, C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4bz7.cif.gz 4bz7.cif.gz | 675.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4bz7.ent.gz pdb4bz7.ent.gz | 560.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4bz7.json.gz 4bz7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bz/4bz7 https://data.pdbj.org/pub/pdb/validation_reports/bz/4bz7 ftp://data.pdbj.org/pub/pdb/validation_reports/bz/4bz7 ftp://data.pdbj.org/pub/pdb/validation_reports/bz/4bz7 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4bz5C  4bz6C  4bz8C  4bz9C  1t67S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 4 molecules ABCD
| #1: Protein | Mass: 50444.875 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-Non-polymers , 5 types, 1205 molecules 

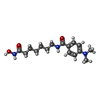






| #2: Chemical | ChemComp-ZN / #3: Chemical | ChemComp-K / #4: Chemical | ChemComp-B3N / #5: Chemical | ChemComp-GOL / #6: Water | ChemComp-HOH / | |
|---|
-Details
| Sequence details | THE GSLVPR MOTIF AT THE END OF THE SEQUENCE CORRESPONDS TO A BAMHI CLONING SITE (GS) FOLLOWED BY A ...THE GSLVPR MOTIF AT THE END OF THE SEQUENCE CORRESPOND |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.5 Å3/Da / Density % sol: 19 % / Description: NONE |
|---|---|
| Crystal grow | Details: 0.2 M NA,K L-TARTRATE, 21% (W/V) PEG3350 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SOLEIL SOLEIL  / Beamline: PROXIMA 1 / Wavelength: 0.9537 / Beamline: PROXIMA 1 / Wavelength: 0.9537 |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Dec 17, 2011 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9537 Å / Relative weight: 1 |
| Reflection | Resolution: 1.65→50 Å / Num. obs: 207485 / % possible obs: 96.9 % / Observed criterion σ(I): 2 / Redundancy: 3.3 % / Biso Wilson estimate: 20.76 Å2 / Rmerge(I) obs: 0.06 / Net I/σ(I): 29.8 |
| Reflection shell | Resolution: 1.65→1.68 Å / Redundancy: 3.3 % / Rmerge(I) obs: 0.21 / Mean I/σ(I) obs: 4.8 / % possible all: 95.5 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1T67 Resolution: 1.65→24.16 Å / Cor.coef. Fo:Fc: 0.9642 / Cor.coef. Fo:Fc free: 0.9598 / SU R Cruickshank DPI: 0.081 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.08 / SU Rfree Blow DPI: 0.075 / SU Rfree Cruickshank DPI: 0.076 Details: IDEAL-DIST CONTACT TERM CONTACT SETUP. RESIDUE TYPES WITHOUT CCP4 ATOM TYPE IN LIBRARY=ZN K. NUMBER OF ATOMS WITH PROPER CCP4 ATOM TYPE=14537. NUMBER WITH APPROX DEFAULT CCP4 ATOM TYPE=0. ...Details: IDEAL-DIST CONTACT TERM CONTACT SETUP. RESIDUE TYPES WITHOUT CCP4 ATOM TYPE IN LIBRARY=ZN K. NUMBER OF ATOMS WITH PROPER CCP4 ATOM TYPE=14537. NUMBER WITH APPROX DEFAULT CCP4 ATOM TYPE=0. NUMBER TREATED BY BAD NON-BONDED CONTACTS=12.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 24.54 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.179 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.65→24.16 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.65→1.69 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj



