+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3rcl | ||||||
|---|---|---|---|---|---|---|---|
| Title | Human Cyclophilin D Complexed with a Fragment | ||||||
 Components Components | Peptidyl-prolyl cis-trans isomerase F, mitochondrial | ||||||
 Keywords Keywords | ISOMERASE/ISOMERASE INHIBITOR / beta barrel / prolyl cis/trans isomerase / mitochondria / inhibitor / ISOMERASE-ISOMERASE INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology information: / : / mitochondrial outer membrane permeabilization involved in programmed cell death / regulation of mitochondrial membrane permeability involved in programmed necrotic cell death / skeletal muscle fiber differentiation / mitochondrial permeability transition pore complex / cellular response to arsenic-containing substance / mitochondrial depolarization / negative regulation of ATP-dependent activity / negative regulation of oxidative phosphorylation ...: / : / mitochondrial outer membrane permeabilization involved in programmed cell death / regulation of mitochondrial membrane permeability involved in programmed necrotic cell death / skeletal muscle fiber differentiation / mitochondrial permeability transition pore complex / cellular response to arsenic-containing substance / mitochondrial depolarization / negative regulation of ATP-dependent activity / negative regulation of oxidative phosphorylation / regulation of mitochondrial membrane permeability / cyclosporin A binding / negative regulation of release of cytochrome c from mitochondria / negative regulation of intrinsic apoptotic signaling pathway / necroptotic process / apoptotic mitochondrial changes / cellular response to calcium ion / response to ischemia / peptidylprolyl isomerase / peptidyl-prolyl cis-trans isomerase activity / cellular response to hydrogen peroxide / protein folding / mitochondrial matrix / negative regulation of apoptotic process / mitochondrion / membrane / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.7 Å MOLECULAR REPLACEMENT / Resolution: 1.7 Å | ||||||
 Authors Authors | Colliandre, L. / Ahmed-Belkacem, H. / Bessin, Y. / Pawlotsky, J.M. / Guichou, J.F. | ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2016 Journal: Nat Commun / Year: 2016Title: Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities. Authors: Ahmed-Belkacem, A. / Colliandre, L. / Ahnou, N. / Nevers, Q. / Gelin, M. / Bessin, Y. / Brillet, R. / Cala, O. / Douguet, D. / Bourguet, W. / Krimm, I. / Pawlotsky, J.M. / Guichou, J.F. | ||||||
| History |
|
- Structure visualization
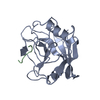
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3rcl.cif.gz 3rcl.cif.gz | 55.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3rcl.ent.gz pdb3rcl.ent.gz | 38.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3rcl.json.gz 3rcl.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rc/3rcl https://data.pdbj.org/pub/pdb/validation_reports/rc/3rcl ftp://data.pdbj.org/pub/pdb/validation_reports/rc/3rcl ftp://data.pdbj.org/pub/pdb/validation_reports/rc/3rcl | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3r49C  3r4gC  3r54C  3r56C  3r57C  3r59C  3rcfC  3rcgC  3rciC  3rckC  3rd9C  3rdaC  3rdbC  3rddC  4j58C  4j59C  4j5bC  4j5cC  4j5dC  4j5eC  2bitS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 17870.400 Da / Num. of mol.: 1 / Fragment: UNP residues 43-207 / Mutation: K175I Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: PPIF, CYP3 / Production host: Homo sapiens (human) / Gene: PPIF, CYP3 / Production host:  |
|---|---|
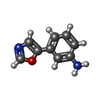
| #2: Chemical | ChemComp-5AO / |
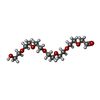
| #3: Chemical | ChemComp-P4C / |
| #4: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.99 Å3/Da / Density % sol: 38.24 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 7.3 Details: 30% PEG4000, pH 7.3, VAPOR DIFFUSION, HANGING DROP, temperature 291K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU300 / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RU300 / Wavelength: 1.5418 |
| Detector | Type: MARRESEARCH / Detector: AREA DETECTOR / Date: Feb 23, 2008 |
| Radiation | Monochromator: yale mirror / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.7→22 Å / Num. all: 15630 / Num. obs: 15333 / % possible obs: 98.1 % / Observed criterion σ(I): 5.9 / Redundancy: 5.9 % / Rsym value: 0.02 |
| Reflection shell | Resolution: 1.7→1.749 Å / Redundancy: 3.7 % / Mean I/σ(I) obs: 3.7 / Rsym value: 0.037 / % possible all: 87.4 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2BIT Resolution: 1.7→21.86 Å / Cor.coef. Fo:Fc: 0.967 / Cor.coef. Fo:Fc free: 0.951 / SU B: 1.579 / SU ML: 0.054 / Cross valid method: THROUGHOUT / ESU R: 0.101 / ESU R Free: 0.097 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 5.838 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.7→21.86 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.705→1.749 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller











 PDBj
PDBj