[English] 日本語
 Yorodumi
Yorodumi- PDB-3obl: Crystal structure of the potent anti-HIV cyanobacterial lectin fr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3obl | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the potent anti-HIV cyanobacterial lectin from Oscillatoria Agardhii | ||||||
 Components Components | Lectin | ||||||
 Keywords Keywords | SUGAR BINDING PROTEIN / novel beta barrel fold / anti-HIV lectin / High mannose glycans | ||||||
| Function / homology | Lipocalin - #450 / OAA-family lectin sugar binding domain / : / OAA-family lectin sugar binding domain / Lipocalin / carbohydrate binding / Beta Barrel / Mainly Beta / Lectin Function and homology information Function and homology information | ||||||
| Biological species |  Planktothrix agardhii (bacteria) Planktothrix agardhii (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 1.2 Å MAD / Resolution: 1.2 Å | ||||||
 Authors Authors | Koharudin, L.M.I. / Furey, W. / Gronenborn, A.M. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2011 Journal: J.Biol.Chem. / Year: 2011Title: Novel fold and carbohydrate specificity of the potent anti-HIV cyanobacterial lectin from Oscillatoria agardhii. Authors: Koharudin, L.M. / Furey, W. / Gronenborn, A.M. #1: Journal: J.Biol.Chem. / Year: 2007 Title: Primary structure and carbohydrate binding specificity of a potent anti-HIV lectin isolated from the filamentous cyanobacterium Oscillatoria agardhii Authors: Sato, Y. / Okuyama, S. / Hori, K. #2:  Journal: Fisheries Sci. / Year: 2009 Journal: Fisheries Sci. / Year: 2009Title: Cloning, expression, and characterization of a novel anti-HIV lectin from the cultured cyanobacterium, Oscillatoria agardhii Authors: Sato, T. / Hori, K. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3obl.cif.gz 3obl.cif.gz | 133.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3obl.ent.gz pdb3obl.ent.gz | 105.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3obl.json.gz 3obl.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ob/3obl https://data.pdbj.org/pub/pdb/validation_reports/ob/3obl ftp://data.pdbj.org/pub/pdb/validation_reports/ob/3obl ftp://data.pdbj.org/pub/pdb/validation_reports/ob/3obl | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 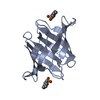
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 13930.758 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: Expressed as native protein / Source: (gene. exp.)  Planktothrix agardhii (bacteria) / Gene: OAA / Plasmid: pET26B(+) / Production host: Planktothrix agardhii (bacteria) / Gene: OAA / Plasmid: pET26B(+) / Production host:  #2: Chemical | ChemComp-CXS / #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.71 Å3/Da / Density % sol: 54.54 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 8 Details: 1.2 M NaH2PO4/0.8 M K2HPO4, 0.2 M Li2SO4, 0.1 M CAPS (pH 10.5), VAPOR DIFFUSION, SITTING DROP, temperature 298K |
-Data collection
| Diffraction | Mean temperature: 277 K | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 22-BM / Wavelength: 0.9795, 0.9793, 0.9718 / Beamline: 22-BM / Wavelength: 0.9795, 0.9793, 0.9718 | ||||||||||||
| Detector | Type: MAR scanner 300 mm plate / Detector: IMAGE PLATE / Date: Feb 25, 2010 / Details: MIRRORS | ||||||||||||
| Radiation | Monochromator: ROSENBAUM-ROCK MONOCHROMATOR HIGH-RESOLUTION DOUBLE-CRYSTAL SI(220) SAGITTAL FOCUSING, ROSENBAUM- ROCK VERTICAL FOCUSING MIRROR Protocol: MAD / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||
| Radiation wavelength |
| ||||||||||||
| Reflection | Resolution: 1.1→22.64 Å / Num. obs: 86175 / % possible obs: 92 % / Observed criterion σ(F): 1 / Observed criterion σ(I): 3 / Redundancy: 3.81 % / Rmerge(I) obs: 0.079 / Net I/σ(I): 7.5 | ||||||||||||
| Reflection shell | Resolution: 1.1→1.14 Å / Redundancy: 3.73 % / Rmerge(I) obs: 0.434 / Mean I/σ(I) obs: 2.1 / Num. unique all: 9892 / % possible all: 83.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MAD / Resolution: 1.2→22.64 Å / Cor.coef. Fo:Fc: 0.976 / Cor.coef. Fo:Fc free: 0.967 / SU B: 1.692 / SU ML: 0.033 / Isotropic thermal model: Anisotropic / Cross valid method: THROUGHOUT / σ(F): 1 / ESU R Free: 0.039 / Stereochemistry target values: MAXIMUM LIKELIHOOD MAD / Resolution: 1.2→22.64 Å / Cor.coef. Fo:Fc: 0.976 / Cor.coef. Fo:Fc free: 0.967 / SU B: 1.692 / SU ML: 0.033 / Isotropic thermal model: Anisotropic / Cross valid method: THROUGHOUT / σ(F): 1 / ESU R Free: 0.039 / Stereochemistry target values: MAXIMUM LIKELIHOODDetails: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. THE DATA HAVE BEEN COLLECTED UP 1.10 A RESOLUTION. HOWEVER, REFINEMENT CONVERGED TO BETTER R VALUES BY CUTTING THE RESOLUTION TO 1.20 A.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 13.854 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.2→22.64 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.2→1.231 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller











 PDBj
PDBj