+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3nd3 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
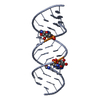
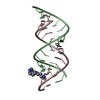
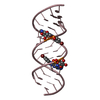
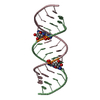
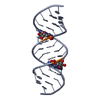
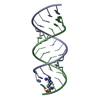
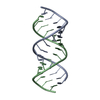
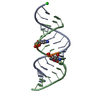
| Title | Uhelix 16-mer dsRNA | ||||||||||||||||||
 Components Components | 5'-R(* Keywords KeywordsRNA / double-stranded RNA / U-helix / oligo-U tail / polyU RNA | Function / homology | : / RNA / RNA (> 10) |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.37 Å molecular replacement / Resolution: 1.37 Å  Authors AuthorsMooers, B.H. / Singh, A. |  Citation Citation Journal: Rna / Year: 2011 Journal: Rna / Year: 2011Title: The crystal structure of an oligo(U):pre-mRNA duplex from a trypanosome RNA editing substrate. Authors: Mooers, B.H. / Singh, A. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3nd3.cif.gz 3nd3.cif.gz | 33.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3nd3.ent.gz pdb3nd3.ent.gz | 24.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3nd3.json.gz 3nd3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nd/3nd3 https://data.pdbj.org/pub/pdb/validation_reports/nd/3nd3 ftp://data.pdbj.org/pub/pdb/validation_reports/nd/3nd3 ftp://data.pdbj.org/pub/pdb/validation_reports/nd/3nd3 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 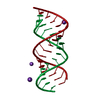
| |||||||||||||||
| Unit cell |
| |||||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: RNA chain | Mass: 5086.016 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: chemically synthesized |
|---|---|
| #2: Chemical | ChemComp-K / |
| #3: Chemical | ChemComp-NA / |
| #4: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.18 Å3/Da / Density % sol: 43.65 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 6 Details: 50 mM NaCacodylate pH 6.0, 10 mM MgCl2, and 1 M LiSO4, vapor diffusion, hanging drop, temperature 298K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL9-2 / Wavelength: 0.9184 Å / Beamline: BL9-2 / Wavelength: 0.9184 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: MARMOSAIC 325 mm CCD / Detector: CCD / Date: Jul 20, 2008 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: GRAPHITE / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.9184 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.37→18.309 Å / Num. all: 9679 / Num. obs: 9679 / % possible obs: 99.8 % / Redundancy: 17.2 % / Rsym value: 0.026 / Net I/σ(I): 52 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.37→18.309 Å / Occupancy max: 1 / Occupancy min: 0.18 / FOM work R set: 0.82 / SU ML: 0.14 / σ(F): 0.48 / Stereochemistry target values: ML MOLECULAR REPLACEMENT / Resolution: 1.37→18.309 Å / Occupancy max: 1 / Occupancy min: 0.18 / FOM work R set: 0.82 / SU ML: 0.14 / σ(F): 0.48 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 44.297 Å2 / ksol: 0.448 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 61.85 Å2 / Biso mean: 34.966 Å2 / Biso min: 19.72 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.37→18.309 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 7
|
 Movie
Movie Controller
Controller





 PDBj
PDBj


































