Entry Database : PDB / ID : 3lp3Title p15 HIV RNaseH domain with inhibitor MK3 p15 Keywords / / / / / / / / / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Method / / Resolution : 2.8 Å Authors Yan, Y. / Munshi, S.K. / Prasad, G.S. / Su, H.P. Journal : J.Virol. / Year : 2010Title : Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.Authors: Su, H.P. / Yan, Y. / Prasad, G.S. / Smith, R.F. / Daniels, C.L. / Abeywickrema, P.D. / Reid, J.C. / Loughran, H.M. / Kornienko, M. / Sharma, S. / Grobler, J.A. / Xu, B. / Sardana, V. / ... Authors : Su, H.P. / Yan, Y. / Prasad, G.S. / Smith, R.F. / Daniels, C.L. / Abeywickrema, P.D. / Reid, J.C. / Loughran, H.M. / Kornienko, M. / Sharma, S. / Grobler, J.A. / Xu, B. / Sardana, V. / Allison, T.J. / Williams, P.D. / Darke, P.L. / Hazuda, D.J. / Munshi, S. History Deposition Feb 4, 2010 Deposition site / Processing site Revision 1.0 Jun 9, 2010 Provider / Type Revision 1.1 Jul 13, 2011 Group Revision 1.2 Nov 1, 2017 Group / Category Item _software.classification / _software.contact_author ... _software.classification / _software.contact_author / _software.contact_author_email / _software.date / _software.language / _software.location / _software.name / _software.type / _software.version Revision 1.3 Feb 21, 2024 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_struct_conn_angle / struct_conn / struct_ncs_dom_lim / struct_ref_seq_dif / struct_site Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id / _struct_ref_seq_dif.details / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 Human immunodeficiency virus type 1
Human immunodeficiency virus type 1 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  FOURIER SYNTHESIS / Resolution: 2.8 Å
FOURIER SYNTHESIS / Resolution: 2.8 Å  Authors
Authors Citation
Citation Journal: J.Virol. / Year: 2010
Journal: J.Virol. / Year: 2010 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 3lp3.cif.gz
3lp3.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb3lp3.ent.gz
pdb3lp3.ent.gz PDB format
PDB format 3lp3.json.gz
3lp3.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/lp/3lp3
https://data.pdbj.org/pub/pdb/validation_reports/lp/3lp3 ftp://data.pdbj.org/pub/pdb/validation_reports/lp/3lp3
ftp://data.pdbj.org/pub/pdb/validation_reports/lp/3lp3 Links
Links Assembly
Assembly
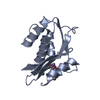


 Components
Components
 Human immunodeficiency virus type 1 / Gene: gag-pol / Production host:
Human immunodeficiency virus type 1 / Gene: gag-pol / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation ROTATING ANODE / Type: RIGAKU FR-E+ SUPERBRIGHT / Wavelength: 1.54 Å
ROTATING ANODE / Type: RIGAKU FR-E+ SUPERBRIGHT / Wavelength: 1.54 Å Processing
Processing FOURIER SYNTHESIS / Resolution: 2.8→34.88 Å / Cor.coef. Fo:Fc: 0.902 / Cor.coef. Fo:Fc free: 0.852 / Occupancy max: 1 / Occupancy min: 1 / SU B: 17.816 / SU ML: 0.346 / Cross valid method: THROUGHOUT / ESU R Free: 0.428 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
FOURIER SYNTHESIS / Resolution: 2.8→34.88 Å / Cor.coef. Fo:Fc: 0.902 / Cor.coef. Fo:Fc free: 0.852 / Occupancy max: 1 / Occupancy min: 1 / SU B: 17.816 / SU ML: 0.346 / Cross valid method: THROUGHOUT / ESU R Free: 0.428 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS Movie
Movie Controller
Controller
















 PDBj
PDBj






