[English] 日本語
 Yorodumi
Yorodumi- PDB-3iai: Crystal structure of the catalytic domain of the tumor-associated... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3iai | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX | |||||||||
 Components Components | Carbonic anhydrase 9 | |||||||||
 Keywords Keywords | LYASE / CARBONIC ANHYDRASES / TRANSMEMBRANE PROTEINS / Cell membrane / Cell projection / Disulfide bond / Glycoprotein / Membrane / Metal-binding / Nucleus / Phosphoprotein / Transmembrane | |||||||||
| Function / homology |  Function and homology information Function and homology informationRegulation of gene expression by Hypoxia-inducible Factor / response to testosterone / microvillus membrane / secretion / Reversible hydration of carbon dioxide / molecular function activator activity / morphogenesis of an epithelium / carbonic anhydrase / carbonate dehydratase activity / basolateral plasma membrane ...Regulation of gene expression by Hypoxia-inducible Factor / response to testosterone / microvillus membrane / secretion / Reversible hydration of carbon dioxide / molecular function activator activity / morphogenesis of an epithelium / carbonic anhydrase / carbonate dehydratase activity / basolateral plasma membrane / response to hypoxia / response to xenobiotic stimulus / nucleolus / zinc ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å | |||||||||
 Authors Authors | Alterio, V. / Di Fiore, A. / De Simone, G. | |||||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2009 Journal: Proc.Natl.Acad.Sci.USA / Year: 2009Title: Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX. Authors: Alterio, V. / Hilvo, M. / Di Fiore, A. / Supuran, C.T. / Pan, P. / Parkkila, S. / Scaloni, A. / Pastorek, J. / Pastorekova, S. / Pedone, C. / Scozzafava, A. / Monti, S.M. / De Simone, G. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3iai.cif.gz 3iai.cif.gz | 261.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3iai.ent.gz pdb3iai.ent.gz | 209.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3iai.json.gz 3iai.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ia/3iai https://data.pdbj.org/pub/pdb/validation_reports/ia/3iai ftp://data.pdbj.org/pub/pdb/validation_reports/ia/3iai ftp://data.pdbj.org/pub/pdb/validation_reports/ia/3iai | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1rj5S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein / Sugars , 2 types, 8 molecules ABCD
| #1: Protein | Mass: 28172.684 Da / Num. of mol.: 4 / Fragment: EXTRACELLULAR CATALYTIC DOMAIN / Mutation: C41S Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CA9, G250, MN / Cell line (production host): Sf9 / Production host: Homo sapiens (human) / Gene: CA9, G250, MN / Cell line (production host): Sf9 / Production host:  #2: Polysaccharide | alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2- ...alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source |
|---|
-Non-polymers , 6 types, 1704 molecules 
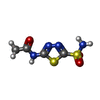









| #3: Chemical | ChemComp-ZN / #4: Chemical | ChemComp-AZM / #5: Chemical | ChemComp-GOL / #6: Chemical | ChemComp-TRS / #7: Chemical | ChemComp-PO4 / #8: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 5.562 Å3/Da / Density % sol: 77.885 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 4 Details: 1.0 M AMMONIUM DIHYDROGEN PHOSPHATE, 0.1 M SODIUM ACETATE, pH 4.0, VAPOR DIFFUSION, HANGING DROP, temperature 298K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ELETTRA ELETTRA  / Beamline: 5.2R / Wavelength: 1 Å / Beamline: 5.2R / Wavelength: 1 Å |
| Detector | Type: MAR CCD 165 mm / Detector: CCD / Date: Feb 14, 2009 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.2→20 Å / Num. all: 122974 / Num. obs: 122974 / % possible obs: 99.3 % / Redundancy: 6.7 % / Rmerge(I) obs: 0.054 / Net I/σ(I): 28.5 |
| Reflection shell | Resolution: 2.2→2.28 Å / Redundancy: 3.8 % / Rmerge(I) obs: 0.231 / Mean I/σ(I) obs: 5.2 / Num. unique all: 11829 / % possible all: 95.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1RJ5 Resolution: 2.2→20 Å / σ(F): 0 / σ(I): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→20 Å
| ||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj







