Entry Database : PDB / ID : 3hs9Title Intersectin 1-peptide-AP2 beta ear complex AP-2 complex subunit beta-1 peptide from Intersectin-1, residues 841-851 Keywords / / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Rattus norvegicus (Norway rat)Method / / / / Resolution : 2.15 Å Authors Vahedi-Faridi, A. / Pechstein, A. / Schaefer, J.G. / Saenger, W. / Haucke, V. Journal : Proc.Natl.Acad.Sci.USA / Year : 2010Title : Regulation of synaptic vesicle recycling by complex formation between intersectin 1 and the clathrin adaptor complex AP2.Authors : Pechstein, A. / Bacetic, J. / Vahedi-Faridi, A. / Gromova, K. / Sundborger, A. / Tomlin, N. / Krainer, G. / Vorontsova, O. / Schafer, J.G. / Owe, S.G. / Cousin, M.A. / Saenger, W. / Shupliakov, O. / Haucke, V. History Deposition Jun 10, 2009 Deposition site / Processing site Revision 1.0 Feb 23, 2010 Provider / Type Revision 1.1 Jul 13, 2011 Group Revision 1.2 Nov 1, 2023 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_ref_seq_dif Item / _database_2.pdbx_database_accession / _struct_ref_seq_dif.details
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.15 Å
molecular replacement / Resolution: 2.15 Å  Authors
Authors Citation
Citation Journal: Proc.Natl.Acad.Sci.USA / Year: 2010
Journal: Proc.Natl.Acad.Sci.USA / Year: 2010 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 3hs9.cif.gz
3hs9.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb3hs9.ent.gz
pdb3hs9.ent.gz PDB format
PDB format 3hs9.json.gz
3hs9.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/hs/3hs9
https://data.pdbj.org/pub/pdb/validation_reports/hs/3hs9 ftp://data.pdbj.org/pub/pdb/validation_reports/hs/3hs9
ftp://data.pdbj.org/pub/pdb/validation_reports/hs/3hs9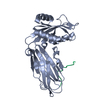

 Links
Links Assembly
Assembly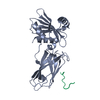
 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  BESSY
BESSY  / Beamline: 14.2 / Wavelength: 0.91841 Å
/ Beamline: 14.2 / Wavelength: 0.91841 Å molecular replacement
molecular replacement Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller













 PDBj
PDBj














