Entry Database : PDB / ID : 3ce4Title Structure of Macrophage Migration Inhibitory Factor Covalently Inhibited by PMSF Treatment Macrophage migration inhibitory factor Keywords / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 1.55 Å Authors Crichlow, G.V. / Lolis, E. Journal : Biochemistry / Year : 2009Title : Structural and kinetic analyses of macrophage migration inhibitory factor active site interactions.Authors : Crichlow, G.V. / Lubetsky, J.B. / Leng, L. / Bucala, R. / Lolis, E.J. History Deposition Feb 28, 2008 Deposition site / Processing site Revision 1.0 Dec 30, 2008 Provider / Type Revision 1.1 Jul 13, 2011 Group / Version format complianceRevision 1.2 Oct 25, 2017 Group / Category Revision 1.3 Aug 30, 2023 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_conn / struct_site Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.pdbx_leaving_atom_flag / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id Revision 1.4 Oct 30, 2024 Group / Category / pdbx_modification_featureRevision 2.0 Apr 23, 2025 Group Advisory / Atomic model ... Advisory / Atomic model / Data collection / Derived calculations / Non-polymer description / Source and taxonomy / Structure summary Category atom_site / chem_comp ... atom_site / chem_comp / chem_comp_atom / chem_comp_bond / entity / entity_name_com / entity_src_gen / pdbx_distant_solvent_atoms / pdbx_entity_nonpoly / pdbx_modification_feature / pdbx_nonpoly_scheme / struct_conn / struct_site Item _atom_site.B_iso_or_equiv / _atom_site.Cartn_x ... _atom_site.B_iso_or_equiv / _atom_site.Cartn_x / _atom_site.Cartn_y / _atom_site.Cartn_z / _atom_site.auth_atom_id / _atom_site.auth_comp_id / _atom_site.label_atom_id / _atom_site.label_comp_id / _atom_site.type_symbol / _chem_comp.formula / _chem_comp.formula_weight / _chem_comp.id / _chem_comp.name / _chem_comp.pdbx_synonyms / _entity.formula_weight / _entity.pdbx_description / _entity.pdbx_ec / _entity_name_com.name / _entity_src_gen.pdbx_beg_seq_num / _entity_src_gen.pdbx_end_seq_num / _entity_src_gen.pdbx_gene_src_ncbi_taxonomy_id / _entity_src_gen.pdbx_host_org_ncbi_taxonomy_id / _entity_src_gen.pdbx_seq_type / _pdbx_entity_nonpoly.comp_id / _pdbx_entity_nonpoly.name / _pdbx_modification_feature.auth_comp_id / _pdbx_modification_feature.label_comp_id / _pdbx_modification_feature.ref_comp_id / _pdbx_modification_feature.ref_pcm_id / _pdbx_nonpoly_scheme.mon_id / _pdbx_nonpoly_scheme.pdb_mon_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_label_comp_id / _struct_site.details / _struct_site.pdbx_auth_comp_id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.55 Å
molecular replacement / Resolution: 1.55 Å  Authors
Authors Citation
Citation Journal: Biochemistry / Year: 2009
Journal: Biochemistry / Year: 2009 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 3ce4.cif.gz
3ce4.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb3ce4.ent.gz
pdb3ce4.ent.gz PDB format
PDB format 3ce4.json.gz
3ce4.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/ce/3ce4
https://data.pdbj.org/pub/pdb/validation_reports/ce/3ce4 ftp://data.pdbj.org/pub/pdb/validation_reports/ce/3ce4
ftp://data.pdbj.org/pub/pdb/validation_reports/ce/3ce4


 Links
Links Assembly
Assembly
 Components
Components Homo sapiens (human) / Gene: MIF, GLIF, MMIF / Plasmid: pET11b / Production host:
Homo sapiens (human) / Gene: MIF, GLIF, MMIF / Plasmid: pET11b / Production host: 









 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418
ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418  molecular replacement
molecular replacement Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller




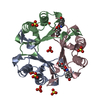
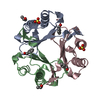





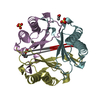
 PDBj
PDBj







