+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3al1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | DESIGNED PEPTIDE ALPHA-1, RACEMIC P1BAR FORM | |||||||||
 Components Components | PROTEIN (D, L-ALPHA-1) | |||||||||
 Keywords Keywords | STRUCTURAL PROTEIN / HELICAL BILAYER / BIOMATERIAL / CENTRIC / RACEMIC | |||||||||
| Function / homology | ETHANOLAMINE Function and homology information Function and homology information | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / DIRECT METHODS / Resolution: 0.75 Å SYNCHROTRON / DIRECT METHODS / Resolution: 0.75 Å | |||||||||
 Authors Authors | Patterson, W.R. / Anderson, D.H. / Degrado, W.F. / Cascio, D. / Eisenberg, D. | |||||||||
 Citation Citation |  Journal: Protein Sci. / Year: 1999 Journal: Protein Sci. / Year: 1999Title: Centrosymmetric bilayers in the 0.75 A resolution structure of a designed alpha-helical peptide, D,L-Alpha-1. Authors: Patterson, W.R. / Anderson, D.H. / DeGrado, W.F. / Cascio, D. / Eisenberg, D. #1:  Journal: To be Published Journal: To be PublishedTitle: Packed Protein Bilayers in the 0.90A Resolution Structure of a Designed Alpha Helical Bundle Authors: Prive, G.G. / Anderson, D.H. / Wesson, L. / Cascio, D. / Eisenberg, D. #2:  Journal: Science / Year: 1990 Journal: Science / Year: 1990Title: Crystal Structure of Alpha-1: Implications for Protein Design Authors: Hill, C.P. / Anderson, D.H. / Wesson, L. / Degrado, W.F. / Eisenberg, D. #3:  Journal: Proteins / Year: 1986 Journal: Proteins / Year: 1986Title: The Design, Synthesis, and Crystallization of an Alpha-Helical Peptide Authors: Eisenberg, D. / Wilcox, W. / Eshita, S.M. / Pryciak, P.M. / Ho, S.P. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3al1.cif.gz 3al1.cif.gz | 36.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3al1.ent.gz pdb3al1.ent.gz | 27.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3al1.json.gz 3al1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/al/3al1 https://data.pdbj.org/pub/pdb/validation_reports/al/3al1 ftp://data.pdbj.org/pub/pdb/validation_reports/al/3al1 ftp://data.pdbj.org/pub/pdb/validation_reports/al/3al1 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 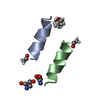
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein/peptide | Mass: 1441.775 Da / Num. of mol.: 2 / Source method: obtained synthetically Details: PEPTIDE WAS SYNTHESIZED VIA SOLID PHASE SYNTHESIS AND DESIGNED TO BE AN AMPHIPHILIC HELIX #2: Chemical | ChemComp-MPD / ( | #3: Chemical | #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.61 Å3/Da / Density % sol: 14.8 % Description: THERE IS NO REDUNDANCY IN THE DATA SET BEYOND 1.28A RESOLUTION; RSYM CAN BE EVALUATED ONLY FOR 18-1.28A. DATA REDUNDANCY IN 18-1.28A SHELL IS 2.51 | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion, hanging drop / pH: 8 Details: EQUAL VOLUMES OF 10 MG/ML D-ALPHA-1 AND 10 MG/ML L-ALPHA-1 WERE MIXED IMMEDIATELY BEFORE CRYSTALLIZATION. THE RESERVOIR SOLUTION CONTAINED 90-93% 2-METHYL-2,4-PENTANEDIOL, 0.075 M ...Details: EQUAL VOLUMES OF 10 MG/ML D-ALPHA-1 AND 10 MG/ML L-ALPHA-1 WERE MIXED IMMEDIATELY BEFORE CRYSTALLIZATION. THE RESERVOIR SOLUTION CONTAINED 90-93% 2-METHYL-2,4-PENTANEDIOL, 0.075 M ETHANOLAMINE HCL PH 9.75, 0.05 M TRIETHANOLAMINE HCL PH 8. THE HANGING DROP CONTAINED EQUAL VOLUMES OF RESERVOIR AND 10 MG/ML D,L MIX ALPHA-1. THE ACIDIC PEPTIDE PARTWAY TITRATES THE BASIC BUFFERS TO REACH THE UNKNOWN RESULTANT PH., vapor diffusion - hanging drop PH range: 8-9.75 | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop / pH: 8 | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 115 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X12C / Wavelength: 0.9 / Beamline: X12C / Wavelength: 0.9 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Jun 1, 1996 |
| Radiation | Monochromator: SI / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9 Å / Relative weight: 1 |
| Reflection | Resolution: 0.75→18 Å / Num. obs: 33573 / % possible obs: 71.5 % / Observed criterion σ(I): 1 / Redundancy: 1.4 % / Rsym value: 10 / Net I/σ(I): 10.6 |
| Reflection shell | Resolution: 0.75→0.78 Å / Redundancy: 1 % / Mean I/σ(I) obs: 2 / % possible all: 24.9 |
| Reflection | *PLUS Rmerge(I) obs: 0.1 |
| Reflection shell | *PLUS % possible obs: 24.9 % / Num. unique obs: 1172 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: DIRECT METHODS / Resolution: 0.75→18 Å / Num. parameters: 2939 / Num. restraintsaints: 4291 / Cross valid method: FREE R / σ(F): 0 StereochEM target val spec case: GLU 108 CONFORMATION C CARBOXYLATE HAS A STIFF PLANARITY RESTRAINT. ETHANOLAMINE AND MPD RESTRAINTS WERE DERIVED BY ANALOGY TO OTHER SIMILAR GROUPS. Stereochemistry target values: ENGH AND HUBER Details: STRUCTURE WAS REDETERMINED WITH SHAKE AND BAKE BECAUSE THE REFINEMENT WAS STUCK AT R ABOUT 21%. THE SNB RESULT CONFIRMED THE SHELXS STRUCTURE. SAME TEST SET WAS USED FOR SHELXL-93 AND -97. ...Details: STRUCTURE WAS REDETERMINED WITH SHAKE AND BAKE BECAUSE THE REFINEMENT WAS STUCK AT R ABOUT 21%. THE SNB RESULT CONFIRMED THE SHELXS STRUCTURE. SAME TEST SET WAS USED FOR SHELXL-93 AND -97. SAME RESTRAINTS WERE CARRIED ALONG INTO SHELXL-97; THERE WAS NO DFIX/DANG DISTINCTION. INTRODUCTION OF ANISOTROPIC REFINEMENT REDUCED FREE R (NO CUTOFF) BY ONLY 1%. IN THIS REFINEMENT, THE ALTERNATE CONFORMATIONS EVENTUALLY REDUCED FREE R FROM 22.6 - 14.5%
| |||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: MOEWS & KRETSINGER, J.MOL.BIOL.91(1973)201-2 | |||||||||||||||||||||||||||||||||
| Refine analyze | Num. disordered residues: 11 / Occupancy sum hydrogen: 250.04 / Occupancy sum non hydrogen: 237.88 | |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 0.75→18 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||
| Software | *PLUS Name: SHELXL-97 / Classification: refinement | |||||||||||||||||||||||||||||||||
| Refinement | *PLUS σ(F): 0 / % reflection Rfree: 10 % / Rfactor obs: 0.13 / Rfactor Rwork: 0.13 | |||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller










 PDBj
PDBj