[English] 日本語
 Yorodumi
Yorodumi- PDB-359d: INHIBITION OF THE HAMMERHEAD RIBOZYME CLEAVAGE REACTION BY SITE-S... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 359d | ||||||
|---|---|---|---|---|---|---|---|
| Title | INHIBITION OF THE HAMMERHEAD RIBOZYME CLEAVAGE REACTION BY SITE-SPECIFIC BINDING OF TB(III) | ||||||
 Components Components | (RNA HAMMERHEAD RIBOZYME) x 2 | ||||||
 Keywords Keywords | RIBOZYME / RNA HAMMERHEAD RIBOZYME / CATALYTIC RNA / LOOP | ||||||
| Function / homology | TERBIUM(III) ION / RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.9 Å SYNCHROTRON / Resolution: 2.9 Å | ||||||
 Authors Authors | Feig, A.L. / Scott, W.G. / Uhlenbeck, O.C. | ||||||
 Citation Citation |  Journal: Science / Year: 1998 Journal: Science / Year: 1998Title: Inhibition of the hammerhead ribozyme cleavage reaction by site-specific binding of Tb. Authors: Feig, A.L. / Scott, W.G. / Uhlenbeck, O.C. #1:  Journal: Science / Year: 1996 Journal: Science / Year: 1996Title: Capturing the Structure of a Catalytic RNA Intermediate: The Hammerhead Ribozyme Authors: Scott, W.G. / Murray, J.B. / Arnold, J.R.P. / Stoddard, B.L. / Klug, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  359d.cif.gz 359d.cif.gz | 30 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb359d.ent.gz pdb359d.ent.gz | 21.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  359d.json.gz 359d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  359d_validation.pdf.gz 359d_validation.pdf.gz | 394.4 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  359d_full_validation.pdf.gz 359d_full_validation.pdf.gz | 401.1 KB | Display | |
| Data in XML |  359d_validation.xml.gz 359d_validation.xml.gz | 4.4 KB | Display | |
| Data in CIF |  359d_validation.cif.gz 359d_validation.cif.gz | 5.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/59/359d https://data.pdbj.org/pub/pdb/validation_reports/59/359d ftp://data.pdbj.org/pub/pdb/validation_reports/59/359d ftp://data.pdbj.org/pub/pdb/validation_reports/59/359d | HTTPS FTP |
-Related structure data
| Related structure data |  299dS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 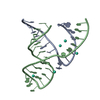
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: RNA chain | Mass: 5170.103 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: RNA chain | Mass: 8019.876 Da / Num. of mol.: 1 / Source method: obtained synthetically |
| #3: Chemical | ChemComp-TB / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 6.49 Å3/Da / Density % sol: 78 % | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion, sitting drop / pH: 6 / Details: pH 6.00, VAPOR DIFFUSION, SITTING DROP | |||||||||||||||||||||||||
| Components of the solutions |
| |||||||||||||||||||||||||
| Crystal | *PLUS | |||||||||||||||||||||||||
| Crystal grow | *PLUS pH: 6 / Method: unknown / Details: Scott, W.G., (1996) Science, 274, 2065. | |||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SRS SRS  / Beamline: PX9.6 / Beamline: PX9.6 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Nov 3, 1996 / Details: MIRRORS |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Relative weight: 1 |
| Reflection | Resolution: 2.9→35.8 Å / Num. obs: 7810 / % possible obs: 97.7 % / Observed criterion σ(I): 0 / Redundancy: 3.9 % / Rmerge(I) obs: 0.052 / Rsym value: 0.047 / Net I/σ(I): 7.7 |
| Reflection shell | Resolution: 2.9→3.06 Å / Redundancy: 4.1 % / Rmerge(I) obs: 0.082 / Mean I/σ(I) obs: 4.8 / Rsym value: 0.079 / % possible all: 100 |
| Reflection | *PLUS Highest resolution: 2.9 Å / Lowest resolution: 35.8 Å / % possible obs: 97.7 % / Observed criterion σ(I): 0 / Redundancy: 3.9 % / Num. measured all: 30459 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Starting model: NDB ENTRY URX057 (PDB ENTRY 299D) Resolution: 2.9→35.8 Å / σ(F): 0 Details: BULK SOLVENT MODEL EMPLOYED TO INCLUDE ALL LOW RESOLUTION DATA
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 52.7 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.9→35.8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file | Serial no: 1 / Param file: NDB_PARAMETER_FILE.RNA / Topol file: NDB_TOPOLOGY_FILE.RNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2.9 Å / Lowest resolution: 35.8 Å / σ(F): 0 / % reflection Rfree: 10 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 52.7 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller





 PDBj
PDBj































