[English] 日本語
 Yorodumi
Yorodumi- PDB-2vzd: Crystal structure of the C-terminal calponin homology domain of a... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2vzd | ||||||
|---|---|---|---|---|---|---|---|
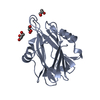
| Title | Crystal structure of the C-terminal calponin homology domain of alpha parvin in complex with paxillin LD1 motif | ||||||
 Components Components |
| ||||||
 Keywords Keywords | CELL ADHESION / CALPONIN HOMOLOGY DOMAIN / CELL MEMBRANE / METAL-BINDING / CYTOSKELETON / CELL JUNCTION / ACTIN-BINDING / PHOSPHOPROTEIN / MEMBRANE / LD1 MOTIF / LIM DOMAIN | ||||||
| Function / homology |  Function and homology information Function and homology informationactin-mediated cell contraction / smooth muscle cell chemotaxis / establishment or maintenance of cell polarity regulating cell shape / Regulation of cytoskeletal remodeling and cell spreading by IPP complex components / Localization of the PINCH-ILK-PARVIN complex to focal adhesions / Regulation of MITF-M-dependent genes involved in extracellular matrix, focal adhesion and epithelial-to-mesenchymal transition / neuropilin binding / vinculin binding / Cell-extracellular matrix interactions / outflow tract septum morphogenesis ...actin-mediated cell contraction / smooth muscle cell chemotaxis / establishment or maintenance of cell polarity regulating cell shape / Regulation of cytoskeletal remodeling and cell spreading by IPP complex components / Localization of the PINCH-ILK-PARVIN complex to focal adhesions / Regulation of MITF-M-dependent genes involved in extracellular matrix, focal adhesion and epithelial-to-mesenchymal transition / neuropilin binding / vinculin binding / Cell-extracellular matrix interactions / outflow tract septum morphogenesis / signal complex assembly / sprouting angiogenesis / microtubule associated complex / heterotypic cell-cell adhesion / growth hormone receptor signaling pathway / establishment or maintenance of cell polarity / protein kinase inhibitor activity / cilium assembly / Smooth Muscle Contraction / GAB1 signalosome / endothelial cell migration / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / stress fiber / positive regulation of stress fiber assembly / substrate adhesion-dependent cell spreading / transforming growth factor beta receptor signaling pathway / cellular response to reactive oxygen species / beta-catenin binding / VEGFA-VEGFR2 Pathway / Z disc / cell-cell junction / cell migration / actin cytoskeleton / lamellipodium / actin binding / actin cytoskeleton organization / cell cortex / protein phosphatase binding / cell adhesion / protein stabilization / cadherin binding / focal adhesion / signal transduction / metal ion binding / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å | ||||||
 Authors Authors | Lorenz, S. / Vakonakis, I. / Lowe, E.D. / Campbell, I.D. / Noble, M.E.M. / Hoellerer, M.K. | ||||||
 Citation Citation |  Journal: Structure / Year: 2008 Journal: Structure / Year: 2008Title: Structural Analysis of the Interactions between Paxillin Ld Motifs and Alpha-Parvin Authors: Lorenz, S. / Vakonakis, I. / Lowe, E.D. / Campbell, I.D. / Noble, M.E.M. / Hoellerer, M.K. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2vzd.cif.gz 2vzd.cif.gz | 82.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2vzd.ent.gz pdb2vzd.ent.gz | 64.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2vzd.json.gz 2vzd.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2vzd_validation.pdf.gz 2vzd_validation.pdf.gz | 495.8 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2vzd_full_validation.pdf.gz 2vzd_full_validation.pdf.gz | 503.1 KB | Display | |
| Data in XML |  2vzd_validation.xml.gz 2vzd_validation.xml.gz | 14.9 KB | Display | |
| Data in CIF |  2vzd_validation.cif.gz 2vzd_validation.cif.gz | 20 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vz/2vzd https://data.pdbj.org/pub/pdb/validation_reports/vz/2vzd ftp://data.pdbj.org/pub/pdb/validation_reports/vz/2vzd ftp://data.pdbj.org/pub/pdb/validation_reports/vz/2vzd | HTTPS FTP |
-Related structure data
| Related structure data |  2vzcSC  2vzgC  2vziC C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Ens-ID: 1 / Refine code: 4
NCS oper: (Code: given Matrix: (-0.2002, 0.867, -0.4563), Vector: |
- Components
Components
-Protein / Protein/peptide , 2 types, 4 molecules ABCD
| #1: Protein | Mass: 15155.380 Da / Num. of mol.: 2 Fragment: C-TERMINAL CALPONIN HOMOLOGY DOMAIN, RESIDUES 242-372 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Plasmid: PGEX-6P1 / Production host: HOMO SAPIENS (human) / Plasmid: PGEX-6P1 / Production host:  #2: Protein/peptide | Mass: 2177.388 Da / Num. of mol.: 2 / Fragment: PAXILLIN LD1 MOTIF, RESIDUES 1-20 / Source method: obtained synthetically / Source: (synth.)  HOMO SAPIENS (human) / References: UniProt: P49023 HOMO SAPIENS (human) / References: UniProt: P49023 |
|---|
-Non-polymers , 5 types, 99 molecules 








| #3: Chemical | | #4: Chemical | #5: Chemical | #6: Chemical | ChemComp-EDO / #7: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.6 Å3/Da / Density % sol: 52.36 % / Description: NONE |
|---|---|
| Crystal grow | pH: 7.5 / Details: 20% (W/V) PEG 10000, 0.1M HEPES PH 7.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID23-1 / Wavelength: 0.969 / Beamline: ID23-1 / Wavelength: 0.969 |
| Detector | Type: ADSC CCD / Detector: CCD / Date: Nov 5, 2006 / Details: MIRRORS |
| Radiation | Monochromator: SI (111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.969 Å / Relative weight: 1 |
| Reflection | Resolution: 2.1→30.03 Å / Num. obs: 20260 / % possible obs: 98.5 % / Observed criterion σ(I): 2 / Redundancy: 2.9 % / Rmerge(I) obs: 0.05 / Net I/σ(I): 16.4 |
| Reflection shell | Resolution: 2.1→2.21 Å / Redundancy: 3 % / Rmerge(I) obs: 0.45 / Mean I/σ(I) obs: 2.5 / % possible all: 99.4 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2VZC Resolution: 2.1→28.64 Å / Cor.coef. Fo:Fc: 0.945 / Cor.coef. Fo:Fc free: 0.923 / SU B: 12.368 / SU ML: 0.169 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / ESU R: 0.294 / ESU R Free: 0.215 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 43.1 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.1→28.64 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj











