+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2nb2 | ||||||
|---|---|---|---|---|---|---|---|
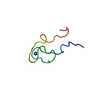
| Title | Nigellin-1.1 | ||||||
 Components Components | nigellin-1.1 | ||||||
 Keywords Keywords | ANTIMICROBIAL PROTEIN / AMP / antimicrobial peptide / plant antimicrobial peptides / defense peptides / blackseed / antifungal activity / antibacterial activity / hairpin-like peptide / alpha-hairpinin | ||||||
| Function / homology | Nigellin-1.1 Function and homology information Function and homology information | ||||||
| Biological species |  Nigella sativa L (black cumin) Nigella sativa L (black cumin) | ||||||
| Method | SOLUTION NMR / simulated annealing, torsion angle dynamics | ||||||
| Model details | fewest violations, model1 | ||||||
 Authors Authors | Bozin, T.N. / Bocharov, E.V. / Rogozhin, E.A. | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: A novel type of hairpin-like defense peptides from blackseed (Nigella sativa L.) contains three disulfide bridges Authors: Rogozhin, E.A. / Kozlov, S.A. / Andreev, Y.A. / Ryazantsev, Y.U. / Grishin, E.V. / Bozin, T.N. / Bocharov, E.V. | ||||||
| History |
|
- Structure visualization
Structure visualization
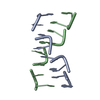
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2nb2.cif.gz 2nb2.cif.gz | 314.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2nb2.ent.gz pdb2nb2.ent.gz | 269.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2nb2.json.gz 2nb2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nb/2nb2 https://data.pdbj.org/pub/pdb/validation_reports/nb/2nb2 ftp://data.pdbj.org/pub/pdb/validation_reports/nb/2nb2 ftp://data.pdbj.org/pub/pdb/validation_reports/nb/2nb2 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 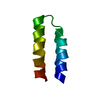
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 4224.847 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Nigella sativa L (black cumin) / References: UniProt: A0A1S4NYD1*PLUS Nigella sativa L (black cumin) / References: UniProt: A0A1S4NYD1*PLUS |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||
| Sample conditions |
|
-NMR measurement
| NMR spectrometer | Type: Varian Uniform NMR System 700 / Manufacturer: Varian / Model: Uniform NMR System 700 / Field strength: 700 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing, torsion angle dynamics / Software ordinal: 1 | |||||||||||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: fewest violations | |||||||||||||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 100 / Conformers submitted total number: 25 |
 Movie
Movie Controller
Controller






 PDBj
PDBj HSQC
HSQC