[English] 日本語
 Yorodumi
Yorodumi- PDB-2mnj: NMR solution structure of the yeast Pih1 and Tah1 C-terminal doma... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2mnj | ||||||
|---|---|---|---|---|---|---|---|
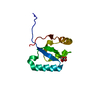
| Title | NMR solution structure of the yeast Pih1 and Tah1 C-terminal domains complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PROTEIN BINDING / CS-domain / R2TP / HSP90 / snoRNP assembly | ||||||
| Function / homology |  Function and homology information Function and homology informationR2TP complex / box C/D snoRNP assembly / regulation of cell size / RNA splicing / mRNA processing / rRNA processing / protein-folding chaperone binding / protein folding / protein stabilization / ribonucleoprotein complex ...R2TP complex / box C/D snoRNP assembly / regulation of cell size / RNA splicing / mRNA processing / rRNA processing / protein-folding chaperone binding / protein folding / protein stabilization / ribonucleoprotein complex / nucleus / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
| Model details | lowest energy, model1 | ||||||
 Authors Authors | Quinternet, M. / Jacquemin, C. / Charpentier, B. / Manival, X. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2015 Journal: J.Mol.Biol. / Year: 2015Title: Structure/Function Analysis of Protein-Protein Interactions Developed by the Yeast Pih1 Platform Protein and Its Partners in Box C/D snoRNP Assembly. Authors: Quinternet, M. / Rothe, B. / Barbier, M. / Bobo, C. / Saliou, J.M. / Jacquemin, C. / Back, R. / Chagot, M.E. / Cianferani, S. / Meyer, P. / Branlant, C. / Charpentier, B. / Manival, X. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2mnj.cif.gz 2mnj.cif.gz | 709.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2mnj.ent.gz pdb2mnj.ent.gz | 596.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2mnj.json.gz 2mnj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mn/2mnj https://data.pdbj.org/pub/pdb/validation_reports/mn/2mnj ftp://data.pdbj.org/pub/pdb/validation_reports/mn/2mnj ftp://data.pdbj.org/pub/pdb/validation_reports/mn/2mnj | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 2555.813 Da / Num. of mol.: 1 / Fragment: UNP residues 93-111 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: TAH1, YCR060W, YCR60W / Plasmid: pnEA-tah1p93-111 / Production host:  |
|---|---|
| #2: Protein | Mass: 10394.039 Da / Num. of mol.: 1 / Fragment: UNP residues 257-344 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: PIH1, NOP17, YHR034C / Plasmid: pnCS-Pih1p257-344 / Production host:  |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 1.5 mM [U-99% 13C; U-99% 15N] Tah1, 1.5 mM [U-99% 13C; U-99% 15N] Pih1, 95% H2O/5% D2O Solvent system: 95% H2O/5% D2O | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||
| Sample conditions | Ionic strength: 50 / pH: 6.4 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Bruker Avance / Manufacturer: Bruker / Model: AVANCE / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 / Details: RECOORD scripts were used | |||||||||||||||
| NMR representative | Selection criteria: lowest energy | |||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller











 PDBj
PDBj



 HSQC
HSQC