+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2mdu | ||||||
|---|---|---|---|---|---|---|---|
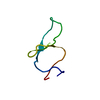
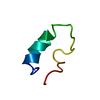
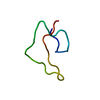
| Title | Circular Permutant of the WW Domain with Loop 1 Excised | ||||||
 Components Components | Pin1 WW domain | ||||||
 Keywords Keywords | ISOMERASE / Circular Permutation / Miniprotein / Dynamics / Folding | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
| Model details | closest to the average, model9 | ||||||
 Authors Authors | Kier, B.L. | ||||||
 Citation Citation |  Journal: J.Am.Chem.Soc. / Year: 2014 Journal: J.Am.Chem.Soc. / Year: 2014Title: Circular Permutation of a WW Domain: Folding Still Occurs after Excising the Turn of the Folding-Nucleating Hairpin. Authors: Kier, B.L. / Anderson, J.M. / Andersen, N.H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2mdu.cif.gz 2mdu.cif.gz | 232.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2mdu.ent.gz pdb2mdu.ent.gz | 193.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2mdu.json.gz 2mdu.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/md/2mdu https://data.pdbj.org/pub/pdb/validation_reports/md/2mdu ftp://data.pdbj.org/pub/pdb/validation_reports/md/2mdu ftp://data.pdbj.org/pub/pdb/validation_reports/md/2mdu | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data | |
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 3794.395 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR Details: This miniprotein construct represents a circular permutation of the Pin1 WW domain (loop 1 excision), with some additional mutations for stability and removal of chromophore redundancy. The ...Details: This miniprotein construct represents a circular permutation of the Pin1 WW domain (loop 1 excision), with some additional mutations for stability and removal of chromophore redundancy. The structure is very similar to that of wild-type WW Pin1, except of course for the termini and loop 1. This miniprotein was not designed with native-like ligand-binding function in mind. | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 50 mM sodium phosphate, 0.5 mM DSS, 90% H2O/10% D2O Solvent system: 90% H2O/10% D2O | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| |||||||||
| Sample conditions | Ionic strength: 0.11 / pH: 6.5 / Pressure: ambient / Temperature: 280 K |
-NMR measurement
| NMR spectrometer | Type: Bruker Avance / Manufacturer: Bruker / Model: AVANCE / Field strength: 700 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 / Details: Standard Lennard-Jones potential | ||||||||||||||||||||
| NMR constraints | NOE constraints total: 473 / NOE intraresidue total count: 222 / NOE long range total count: 118 / NOE medium range total count: 32 / NOE sequential total count: 101 | ||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 40 / Conformers submitted total number: 22 |
 Movie
Movie Controller
Controller












 PDBj
PDBj
