[English] 日本語
 Yorodumi
Yorodumi- PDB-2lpt: Molecular dynamics re-refinement of domain 5 of the Pylaiella lit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2lpt | ||||||
|---|---|---|---|---|---|---|---|
| Title | Molecular dynamics re-refinement of domain 5 of the Pylaiella littoralis group II intron | ||||||
 Components Components | RNA_(34-MER) | ||||||
 Keywords Keywords | RNA / Tetraloop / Bulge | ||||||
| Function / homology | RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / molecular dynamics | ||||||
| Model details | closest to the average, model 3 | ||||||
 Authors Authors | Henriksen, N.M. / Davis, D.R. / Cheatham III, T.E. | ||||||
 Citation Citation |  Journal: Rna / Year: 2006 Journal: Rna / Year: 2006Title: Structure of a self-splicing group II intron catalytic effector domain 5: parallels with spliceosomal U6 RNA. Authors: Seetharaman, M. / Eldho, N.V. / Padgett, R.A. / Dayie, K.T. | ||||||
| History |
| ||||||
| Remark 0 | THIS ENTRY 2LPT REFLECTS AN ALTERNATIVE MODELING OF THE ORIGINAL STRUCTURAL DATA (2F88.MR) ...THIS ENTRY 2LPT REFLECTS AN ALTERNATIVE MODELING OF THE ORIGINAL STRUCTURAL DATA (2F88.MR) DETERMINED BY AUTHORS OF THE PDB ENTRY 2F88: M.SEETHARAMAN,N.V.ELDHO,R.A.PADGETT,K.T.DAYIE |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2lpt.cif.gz 2lpt.cif.gz | 114 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2lpt.ent.gz pdb2lpt.ent.gz | 91.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2lpt.json.gz 2lpt.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lp/2lpt https://data.pdbj.org/pub/pdb/validation_reports/lp/2lpt ftp://data.pdbj.org/pub/pdb/validation_reports/lp/2lpt ftp://data.pdbj.org/pub/pdb/validation_reports/lp/2lpt | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 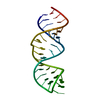
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 11009.607 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR Details: This is a Molecular Dynamics re-refinement of structures from the PDB: 2F88 ensemble using the NMR restraints. The re-refinement shows significant changes in the overall geometry. |
|---|---|
| NMR details | Text: AUTHOR USED THE MR DATA FROM ENTRY 2F88. |
- Processing
Processing
| NMR software | Name:  Amber / Version: 11 Amber / Version: 11 Developer: Case, Darden, Cheatham, III, Simmerling, Wang, Duke, Luo, and Kollman Classification: refinement |
|---|---|
| Refinement | Method: molecular dynamics / Software ordinal: 1 |
| NMR representative | Selection criteria: closest to the average |
| NMR ensemble | Conformer selection criteria: all calculated structures submitted Conformers calculated total number: 5 / Conformers submitted total number: 5 |
 Movie
Movie Controller
Controller







 PDBj
PDBj





























