+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2l3e | ||||||
|---|---|---|---|---|---|---|---|
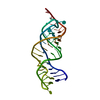
| Title | Solution structure of P2a-J2a/b-P2b of human telomerase RNA | ||||||
 Components Components | 35-MER | ||||||
 Keywords Keywords | RNA / human telomerase RNA / hTR / P2a / P2b / J2ab | ||||||
| Function / homology | RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
| Model details | lowest energy, model 1 | ||||||
 Authors Authors | Zhang, Q. / Kim, N. / Peterson, R.D. / Wang, Z. / Feigon, J. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2010 Journal: Proc.Natl.Acad.Sci.USA / Year: 2010Title: Inaugural Article: Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Authors: Zhang, Q. / Kim, N.K. / Peterson, R.D. / Wang, Z. / Feigon, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2l3e.cif.gz 2l3e.cif.gz | 428.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2l3e.ent.gz pdb2l3e.ent.gz | 359.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2l3e.json.gz 2l3e.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/l3/2l3e https://data.pdbj.org/pub/pdb/validation_reports/l3/2l3e ftp://data.pdbj.org/pub/pdb/validation_reports/l3/2l3e ftp://data.pdbj.org/pub/pdb/validation_reports/l3/2l3e | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 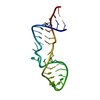
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 11164.637 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||||||||||||||||||
| Sample conditions |
|
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 | ||||||||||||||||||||||||||||||||||||
| NMR constraints | NA alpha-angle constraints total count: 27 / NA beta-angle constraints total count: 25 / NA chi-angle constraints total count: 35 / NA delta-angle constraints total count: 35 / NA epsilon-angle constraints total count: 29 / NA gamma-angle constraints total count: 26 / NA other-angle constraints total count: 53 / NA sugar pucker constraints total count: 35 | ||||||||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||||||||||
| NMR ensemble | Average torsion angle constraint violation: 0.041 ° Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 200 / Conformers submitted total number: 20 / Maximum lower distance constraint violation: 0.08 Å / Maximum torsion angle constraint violation: 1.704 ° / Maximum upper distance constraint violation: 0.412 Å / Torsion angle constraint violation method: X-PLOR NIH | ||||||||||||||||||||||||||||||||||||
| NMR ensemble rms | Distance rms dev: 0.018 Å / Distance rms dev error: 0.003 Å |
 Movie
Movie Controller
Controller






 PDBj
PDBj





























 HSQC
HSQC