| Entry | Database: PDB / ID: 2l0b
|
|---|
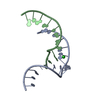
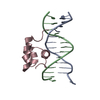
| Title | Solution NMR structure of zinc finger domain of E3 ubiquitin-protein ligase praja-1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) target HR4710B |
|---|
 Components Components | E3 ubiquitin-protein ligase Praja-1 |
|---|
 Keywords Keywords | LIGASE / Zinc finger / NESG / Structural Genomics / PSI-2 / Protein Structure Initiative / Northeast Structural Genomics Consortium |
|---|
| Function / homology |  Function and homology information Function and homology information
protein catabolic process / RING-type E3 ubiquitin transferase / ubiquitin protein ligase activity / Antigen processing: Ubiquitination & Proteasome degradation / protein ubiquitination / zinc ion binding / cytoplasmSimilarity search - Function Ring finger domain / Zinc/RING finger domain, C3HC4 (zinc finger) / Herpes Virus-1 / Ring finger / Zinc finger RING-type profile. / Zinc finger, RING-type / Zinc finger, RING/FYVE/PHD-type / 2-Layer Sandwich / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
|---|
| Method | SOLUTION NMR / distance geometry, simulated annealing, molecular dynamics |
|---|
| Model details | lowest energy, model 1 |
|---|
 Authors Authors | Liu, G. / Tong, S. / Hamilton, K. / Ciccosanti, C. / Shastry, R. / Acton, T.B. / Xiao, R. / Everett, J.K. / Montelione, G.T. / Northeast Structural Genomics Consortium (NESG) |
|---|
 Citation Citation |  Journal: To be Published Journal: To be Published
Title: Solution structure of zinc finger domain of E3 ubiquitin-protein ligase protein praja-1 from Homo sapiens, northeast structural genomics consortium (NESG) target HR4710B
Authors: Liu, G. / Tong, S. / Xiao, R. / Acton, T.B. / Everett, J.K. / Montelione, G.T. |
|---|
| History | | Deposition | Jun 30, 2010 | Deposition site: BMRB / Processing site: RCSB |
|---|
| Revision 1.0 | Aug 25, 2010 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jul 13, 2011 | Group: Version format compliance |
|---|
| Revision 1.2 | May 1, 2024 | Group: Data collection / Database references / Derived calculations
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_nmr_software / pdbx_nmr_spectrometer / pdbx_struct_conn_angle / struct_conn / struct_ref_seq_dif / struct_site
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_nmr_software.name / _pdbx_nmr_spectrometer.model / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_ref_seq_dif.details / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) Authors
Authors Citation
Citation Journal: To be Published
Journal: To be Published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2l0b.cif.gz
2l0b.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2l0b.ent.gz
pdb2l0b.ent.gz PDB format
PDB format 2l0b.json.gz
2l0b.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/l0/2l0b
https://data.pdbj.org/pub/pdb/validation_reports/l0/2l0b ftp://data.pdbj.org/pub/pdb/validation_reports/l0/2l0b
ftp://data.pdbj.org/pub/pdb/validation_reports/l0/2l0b Links
Links Assembly
Assembly
 Components
Components Homo sapiens (human) / Gene: PJA1, RNF70 / Production host:
Homo sapiens (human) / Gene: PJA1, RNF70 / Production host: 
 Sample preparation
Sample preparation Processing
Processing Movie
Movie Controller
Controller








 PDBj
PDBj




 HSQC
HSQC