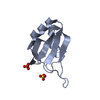
Entry Database : PDB / ID : 2ku7Title Solution structure of MLL1 PHD3-Cyp33 RRM chimeric protein MLL1 PHD3-Cyp33 RRM chimeric protein Keywords / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / Authors Song, J. / Wang, Z. / Patel, D. Journal : Cell(Cambridge,Mass.) / Year : 2010Title : Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression.Authors : Wang, Z. / Song, J. / Milne, T.A. / Wang, G.G. / Li, H. / Allis, C.D. / Patel, D.J. History Deposition Feb 12, 2010 Deposition site / Processing site Revision 1.0 Jul 7, 2010 Provider / Type Revision 1.1 Jul 13, 2011 Group Revision 1.2 Mar 16, 2022 Group / Database references / Derived calculationsCategory database_2 / pdbx_nmr_software ... database_2 / pdbx_nmr_software / pdbx_nmr_spectrometer / pdbx_struct_assembly / pdbx_struct_oper_list / struct_ref_seq_dif Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_nmr_software.name / _pdbx_nmr_spectrometer.model / _struct_ref_seq_dif.details Revision 1.3 May 22, 2024 Group / Category / chem_comp_bond
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) Authors
Authors Citation
Citation Journal: Cell(Cambridge,Mass.) / Year: 2010
Journal: Cell(Cambridge,Mass.) / Year: 2010 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2ku7.cif.gz
2ku7.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2ku7.ent.gz
pdb2ku7.ent.gz PDB format
PDB format 2ku7.json.gz
2ku7.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/ku/2ku7
https://data.pdbj.org/pub/pdb/validation_reports/ku/2ku7 ftp://data.pdbj.org/pub/pdb/validation_reports/ku/2ku7
ftp://data.pdbj.org/pub/pdb/validation_reports/ku/2ku7 Links
Links Assembly
Assembly
 Components
Components Homo sapiens (human) / Plasmid: pRSFDuet-1 / Production host:
Homo sapiens (human) / Plasmid: pRSFDuet-1 / Production host: 
 Sample preparation
Sample preparation Processing
Processing Movie
Movie Controller
Controller














 PDBj
PDBj




 HSQC
HSQC