| 登録情報 | データベース: PDB / ID: 2kri
|
|---|
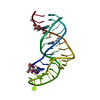
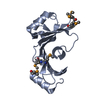
| タイトル | Structure of a complex between domain V of beta2-glycoprotein I and the fourth ligand-binding module from LDLR determined with Haddock |
|---|
 要素 要素 | - Beta-2-glycoprotein 1
- Low-density lipoprotein receptor
|
|---|
 キーワード キーワード | PROTEIN BINDING/ENDOCYTOSIS / Antiphospholipid syndrome / Thrombosis / LDLR / Receptor / Disulfide bond / Glycoprotein / Heparin-binding / Sushi / PROTEIN BINDING-ENDOCYTOSIS complex |
|---|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報
positive regulation of triglyceride metabolic process / triglyceride transport / lipoprotein lipase activator activity / chylomicron remodeling / platelet dense granule lumen / receptor-mediated endocytosis involved in cholesterol transport / regulation of phosphatidylcholine catabolic process / plasma lipoprotein particle clearance / positive regulation of lysosomal protein catabolic process / lipase binding ...positive regulation of triglyceride metabolic process / triglyceride transport / lipoprotein lipase activator activity / chylomicron remodeling / platelet dense granule lumen / receptor-mediated endocytosis involved in cholesterol transport / regulation of phosphatidylcholine catabolic process / plasma lipoprotein particle clearance / positive regulation of lysosomal protein catabolic process / lipase binding / negative regulation of astrocyte activation / negative regulation of microglial cell activation / very-low-density lipoprotein particle receptor activity / PCSK9-LDLR complex / cholesterol import / low-density lipoprotein particle clearance / positive regulation of triglyceride biosynthetic process / clathrin heavy chain binding / negative regulation of receptor recycling / intestinal cholesterol absorption / blood coagulation, intrinsic pathway / low-density lipoprotein particle receptor activity / very-low-density lipoprotein particle remodeling / Chylomicron clearance / amyloid-beta clearance by cellular catabolic process / low-density lipoprotein particle binding / response to caloric restriction / LDL clearance / high-density lipoprotein particle clearance / regulation of fibrinolysis / negative regulation of myeloid cell apoptotic process / chylomicron / regulation of protein metabolic process / lipoprotein catabolic process / phospholipid transport / low-density lipoprotein particle / cholesterol transport / high-density lipoprotein particle / very-low-density lipoprotein particle / cellular response to fatty acid / negative regulation of amyloid fibril formation / endolysosome membrane / negative regulation of low-density lipoprotein particle clearance / negative regulation of endothelial cell migration / regulation of cholesterol metabolic process / artery morphogenesis / negative regulation of protein metabolic process / plasminogen activation / sorting endosome / negative regulation of endothelial cell proliferation / amyloid-beta clearance / negative regulation of smooth muscle cell apoptotic process / lipoprotein particle binding / cellular response to low-density lipoprotein particle stimulus / negative regulation of blood coagulation / positive regulation of blood coagulation / negative regulation of fibrinolysis / long-term memory / phagocytosis / retinoid metabolic process / cholesterol metabolic process / Retinoid metabolism and transport / clathrin-coated pit / somatodendritic compartment / receptor-mediated endocytosis / cholesterol homeostasis / negative regulation of angiogenesis / clathrin-coated endocytic vesicle membrane / phospholipid binding / lipid metabolic process / endocytosis / apical part of cell / positive regulation of inflammatory response / late endosome / Platelet degranulation / Cargo recognition for clathrin-mediated endocytosis / heparin binding / Clathrin-mediated endocytosis / : / amyloid-beta binding / virus receptor activity / protease binding / basolateral plasma membrane / molecular adaptor activity / early endosome / lysosome / receptor complex / endosome membrane / negative regulation of gene expression / external side of plasma membrane / intracellular membrane-bounded organelle / calcium ion binding / lipid binding / positive regulation of gene expression / cell surface / Golgi apparatus / extracellular space / extracellular exosome / extracellular region / identical protein binding類似検索 - 分子機能 Beta-2-glycoprotein-1 fifth domain / Beta-2-glycoprotein-1 fifth domain / : / Complement Module, domain 1 / Complement Module; domain 1 / Low-density lipoprotein receptor repeat class B / LDL-receptor class B (LDLRB) repeat profile. / LDLR class B repeat / Low-density lipoprotein-receptor YWTD domain / Low-density lipoprotein receptor domain class A ...Beta-2-glycoprotein-1 fifth domain / Beta-2-glycoprotein-1 fifth domain / : / Complement Module, domain 1 / Complement Module; domain 1 / Low-density lipoprotein receptor repeat class B / LDL-receptor class B (LDLRB) repeat profile. / LDLR class B repeat / Low-density lipoprotein-receptor YWTD domain / Low-density lipoprotein receptor domain class A / : / Calcium-binding EGF domain / Low-density lipoprotein (LDL) receptor class A, conserved site / LDL-receptor class A (LDLRA) domain signature. / Sushi repeat (SCR repeat) / LDL-receptor class A (LDLRA) domain profile. / Domain abundant in complement control proteins; SUSHI repeat; short complement-like repeat (SCR) / Sushi/SCR/CCP domain / Sushi/CCP/SCR domain profile. / Sushi/SCR/CCP superfamily / Low-density lipoprotein receptor domain class A / Low-density lipoprotein (LDL) receptor class A repeat / LDL receptor-like superfamily / Six-bladed beta-propeller, TolB-like / Coagulation Factor Xa inhibitory site / EGF-type aspartate/asparagine hydroxylation site / EGF-like calcium-binding, conserved site / Calcium-binding EGF-like domain signature. / Aspartic acid and asparagine hydroxylation site. / EGF-like calcium-binding domain / Calcium-binding EGF-like domain / Epidermal growth factor-like domain. / EGF-like domain profile. / Growth factor receptor cysteine-rich domain superfamily / EGF-like domain signature 2. / EGF-like domain / Ribbon / Mainly Beta類似検索 - ドメイン・相同性 Low-density lipoprotein receptor / Beta-2-glycoprotein 1類似検索 - 構成要素 |
|---|
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) |
|---|
| 手法 | 溶液NMR / simulated annealing |
|---|
| Model details | lowest haddock score, model 1 |
|---|
 データ登録者 データ登録者 | Beglova, N. |
|---|
 引用 引用 |  ジャーナル: Structure / 年: 2010 ジャーナル: Structure / 年: 2010
タイトル: Mode of interaction between beta2GPI and lipoprotein receptors suggests mutually exclusive binding of beta2GPI to the receptors and anionic phospholipids.
著者: Lee, C.J. / De Biasio, A. / Beglova, N. |
|---|
| 履歴 | | 登録 | 2009年12月18日 | 登録サイト: BMRB / 処理サイト: RCSB |
|---|
| 改定 1.0 | 2010年3月31日 | Provider: repository / タイプ: Initial release |
|---|
| 改定 1.1 | 2011年7月13日 | Group: Version format compliance |
|---|
| 改定 1.2 | 2019年11月6日 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Experimental preparation
カテゴリ: pdbx_database_related / pdbx_nmr_sample_details ...pdbx_database_related / pdbx_nmr_sample_details / pdbx_nmr_software / pdbx_struct_assembly / pdbx_struct_assembly_prop / pdbx_struct_oper_list / struct_ref_seq_dif
Item: _pdbx_database_related.db_id / _pdbx_nmr_sample_details.contents ..._pdbx_database_related.db_id / _pdbx_nmr_sample_details.contents / _pdbx_nmr_software.name / _struct_ref_seq_dif.details |
|---|
| 改定 1.3 | 2024年10月16日 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Structure summary
カテゴリ: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_entry_details / pdbx_modification_feature / pdbx_struct_conn_angle / struct_conn / struct_site
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id |
|---|
|
|---|
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) データ登録者
データ登録者 引用
引用 ジャーナル: Structure / 年: 2010
ジャーナル: Structure / 年: 2010 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 2kri.cif.gz
2kri.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb2kri.ent.gz
pdb2kri.ent.gz PDB形式
PDB形式 2kri.json.gz
2kri.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 2kri_validation.pdf.gz
2kri_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 2kri_full_validation.pdf.gz
2kri_full_validation.pdf.gz 2kri_validation.xml.gz
2kri_validation.xml.gz 2kri_validation.cif.gz
2kri_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/kr/2kri
https://data.pdbj.org/pub/pdb/validation_reports/kr/2kri ftp://data.pdbj.org/pub/pdb/validation_reports/kr/2kri
ftp://data.pdbj.org/pub/pdb/validation_reports/kr/2kri リンク
リンク 集合体
集合体
 要素
要素 Homo sapiens (ヒト) / 遺伝子: APOH, B2G1 / 発現宿主:
Homo sapiens (ヒト) / 遺伝子: APOH, B2G1 / 発現宿主: 
 Homo sapiens (ヒト) / 遺伝子: LDLR / 発現宿主:
Homo sapiens (ヒト) / 遺伝子: LDLR / 発現宿主: 
 試料調製
試料調製 解析
解析 ムービー
ムービー コントローラー
コントローラー









 PDBj
PDBj















 HSQC
HSQC