[English] 日本語
 Yorodumi
Yorodumi- PDB-2ieo: Crystal structure analysis of HIV-1 protease mutant I84V with a p... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2ieo | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure analysis of HIV-1 protease mutant I84V with a potent non-peptide inhibitor (UIC-94017) | |||||||||
 Components Components | Protease | |||||||||
 Keywords Keywords | HYDROLASE / HIV-1 protease / mutant / I84V / dimer / inhibitor / UIC-94017 / TMC114 / darunavir | |||||||||
| Function / homology |  Function and homology information Function and homology informationHIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / RNA-directed DNA polymerase / establishment of integrated proviral latency / RNA stem-loop binding ...HIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / RNA-directed DNA polymerase / establishment of integrated proviral latency / RNA stem-loop binding / viral penetration into host nucleus / host multivesicular body / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / host cell / viral nucleocapsid / DNA recombination / DNA-directed DNA polymerase / aspartic-type endopeptidase activity / Hydrolases; Acting on ester bonds / DNA-directed DNA polymerase activity / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont entry into host cell / lipid binding / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / DNA binding / zinc ion binding / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.53 Å MOLECULAR REPLACEMENT / Resolution: 1.53 Å | |||||||||
 Authors Authors | Tie, Y. / Boross, P.I. / Wang, Y.F. / Gaddis, L. / Manna, D. / Hussain, A.K. / Leshchenko, S. / Ghosh, A.K. / Louis, J.M. / Harrison, R.W. / Weber, I.T. | |||||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2004 Journal: J.Mol.Biol. / Year: 2004Title: High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-Peptide Inhibitor (Uic-94017) Active Against Multi-Drug-Resistant Clinical Strains. Authors: Tie, Y. / Boross, P.I. / Wang, Y.F. / Gaddis, L. / Hussain, A.K. / Leshchenko, S. / Ghosh, A.K. / Louis, J.M. / Harrison, R.W. / Weber, I.T. | |||||||||
| History |
|
- Structure visualization
Structure visualization
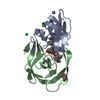
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2ieo.cif.gz 2ieo.cif.gz | 114.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2ieo.ent.gz pdb2ieo.ent.gz | 87.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2ieo.json.gz 2ieo.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2ieo_validation.pdf.gz 2ieo_validation.pdf.gz | 1.1 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2ieo_full_validation.pdf.gz 2ieo_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  2ieo_validation.xml.gz 2ieo_validation.xml.gz | 15.4 KB | Display | |
| Data in CIF |  2ieo_validation.cif.gz 2ieo_validation.cif.gz | 21.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ie/2ieo https://data.pdbj.org/pub/pdb/validation_reports/ie/2ieo ftp://data.pdbj.org/pub/pdb/validation_reports/ie/2ieo ftp://data.pdbj.org/pub/pdb/validation_reports/ie/2ieo | HTTPS FTP |
-Related structure data
| Related structure data |  2idwC 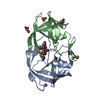 2ienC  1s6g C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 10726.650 Da / Num. of mol.: 2 / Fragment: residues 500-598 Mutation: Q7K, L33I, L63I, C67A, I84V, C95A, Q107K, L133I, L163I, C167A, C195A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Human immunodeficiency virus 1 / Genus: Lentivirus / Gene: gag, pol / Plasmid: PET11A / Species (production host): Escherichia coli / Production host: Human immunodeficiency virus 1 / Genus: Lentivirus / Gene: gag, pol / Plasmid: PET11A / Species (production host): Escherichia coli / Production host:  References: UniProt: P03368, UniProt: Q7SSI0*PLUS, HIV-1 retropepsin #2: Chemical | #3: Chemical | ChemComp-NA / | #4: Chemical | ChemComp-017 / ( | #5: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.62 Å3/Da / Density % sol: 52.68 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 5 Details: SODIUM CHLORIDE, CITRATE PHOSPHATE, DMSO, pH 5.00, VAPOR DIFFUSION, HANGING DROP, temperature 298K |
-Data collection
| Diffraction | Mean temperature: 90 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 22-ID / Wavelength: 1 / Beamline: 22-ID / Wavelength: 1 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Dec 12, 2002 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.53→50 Å / Num. obs: 35966 / % possible obs: 99.3 % / Observed criterion σ(I): 3 / Redundancy: 9.4 % / Rmerge(I) obs: 0.082 / Net I/σ(I): 17.4 |
| Reflection shell | Resolution: 1.53→1.58 Å / Redundancy: 6.2 % / Rmerge(I) obs: 0.312 / % possible all: 97.3 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1S6G  1s6g Resolution: 1.53→10 Å / Num. parameters: 18230 / Num. restraintsaints: 23389 / Stereochemistry target values: Engh & Huber
| |||||||||||||||||||||||||||||||||
| Refine analyze | Num. disordered residues: 15 / Occupancy sum hydrogen: 1628 / Occupancy sum non hydrogen: 1740 | |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.53→10 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj